The first whole genome shotgun assemblies of microorganisms were published in 1995. Since then over 313,000 genomes have been sequenced to varying degrees.
DNA sequencing has become the new framework upon which the Tree of Life is now built and referenced. 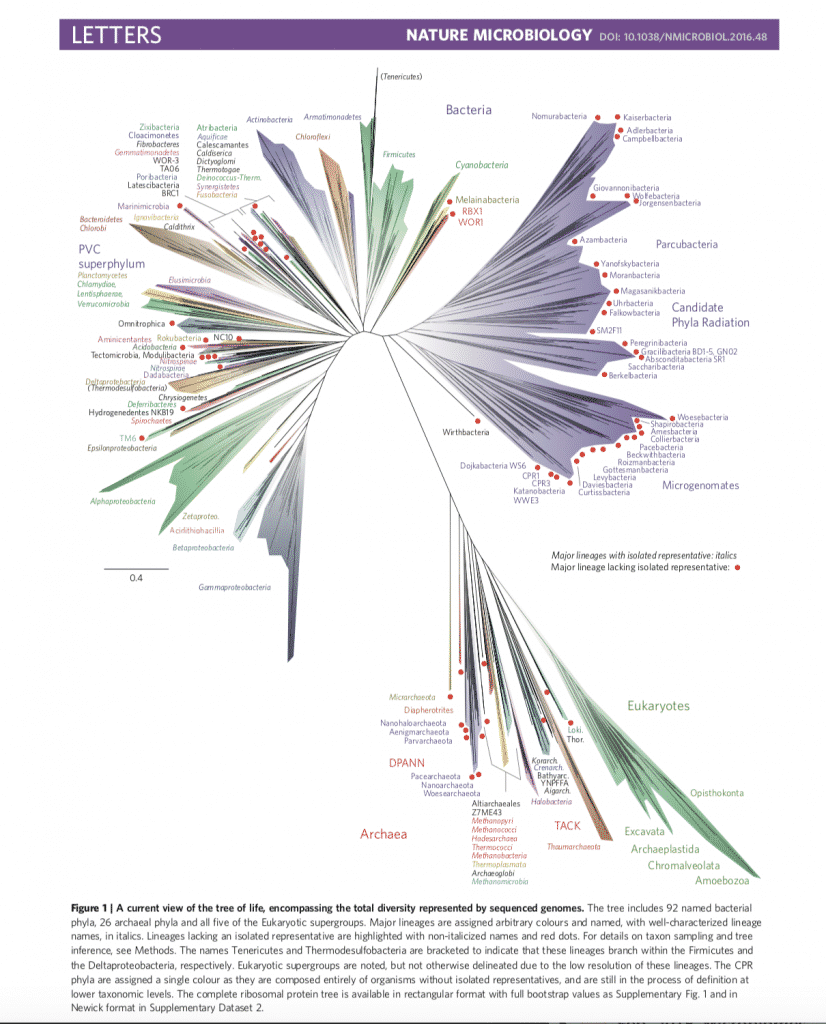
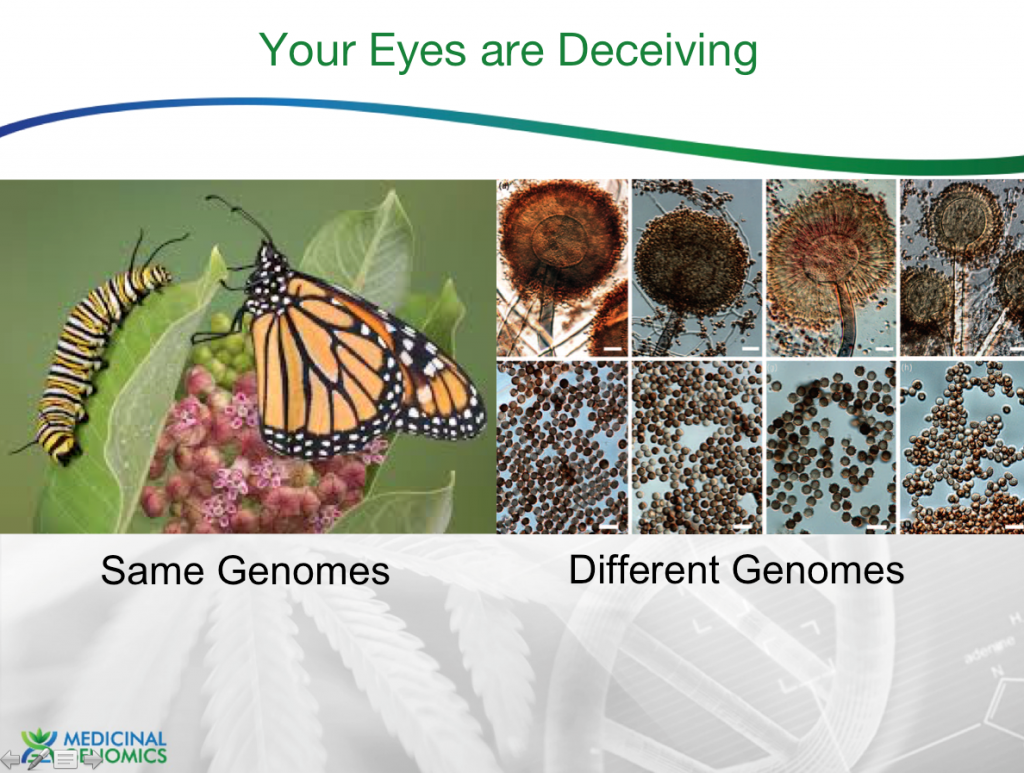
Traditional taxonomy relied on phenotypic features, interbreeding characteristics and metabolic tests. The visual phenotyping has proven to be particularly error prone given the phenotypic diversity some organisms can display over the course of their lifecycle.
The classic example is the Monarch butterfly which undergoes substantial metamorphosis through its lifecycle but relies on the same genome. The flip side of this problem is the case with Aspergillus identification. Only one of the Aspergillus species above is listed as a pathogenic microbe for cannabis testing. The others are often mistaken for it. 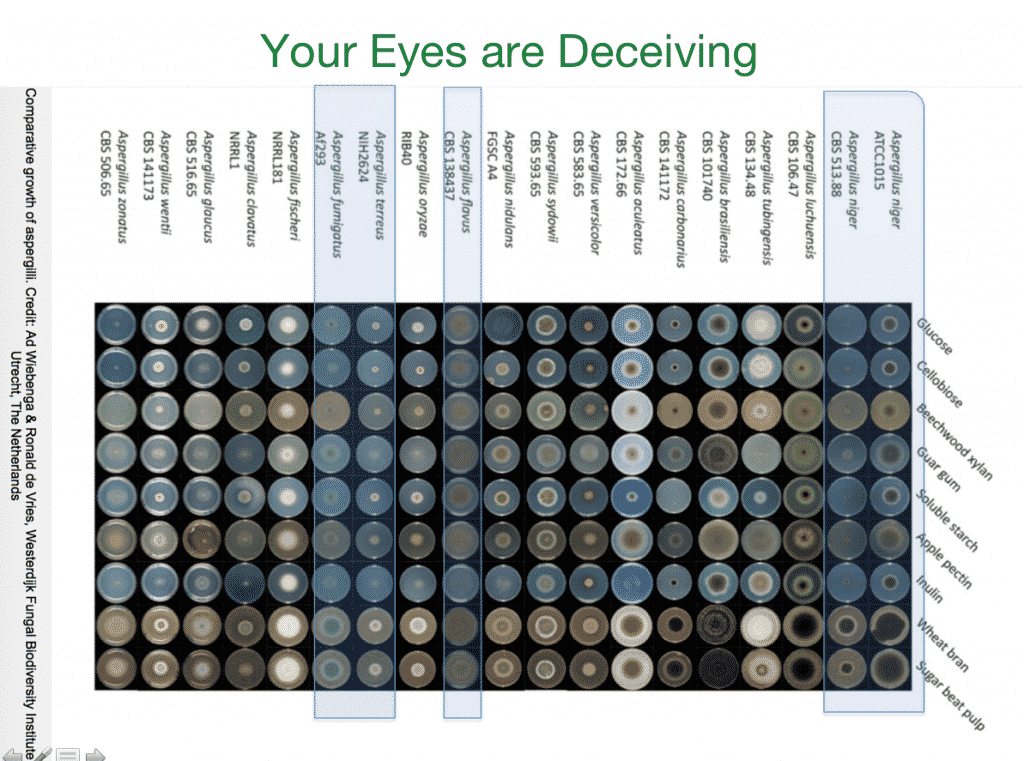
Above are 18 different Aspergillus species plated on 9 different medias. One can see how challenging it can be to visually identify the 4 Aspergillus (terreus, niger, flavus, fumigatus) species in the California cannabis regulations.
For this reason, if there ever is disagreement regarding an Aspergillus qPCR result, we recommend you send the sample in for DNA sequencing. Sending the sample off to be plated is unlikely to resolve the identification.
ITS sequencing can be performed to identify the Aspergillus to the genus level with short read Illumina sequencing. Species level classification may require whole genome shotgun sequencing. This is more easily performed on Aspergillus that has been cultured as it will enrich the Aspergillus DNA above background Cannabis DNA levels. It is important to note that the microbiome that exists directly on the cannabis flower rarely reflects the microbiome seen after culturing. This “Heisenberg uncertainty effect” is widely recognized in the metagenomics field and has been published to occur on Cannabis.
Ivan Lianchko from Phase Genomics underscores this point 2 minutes into his discussion on HiC data.
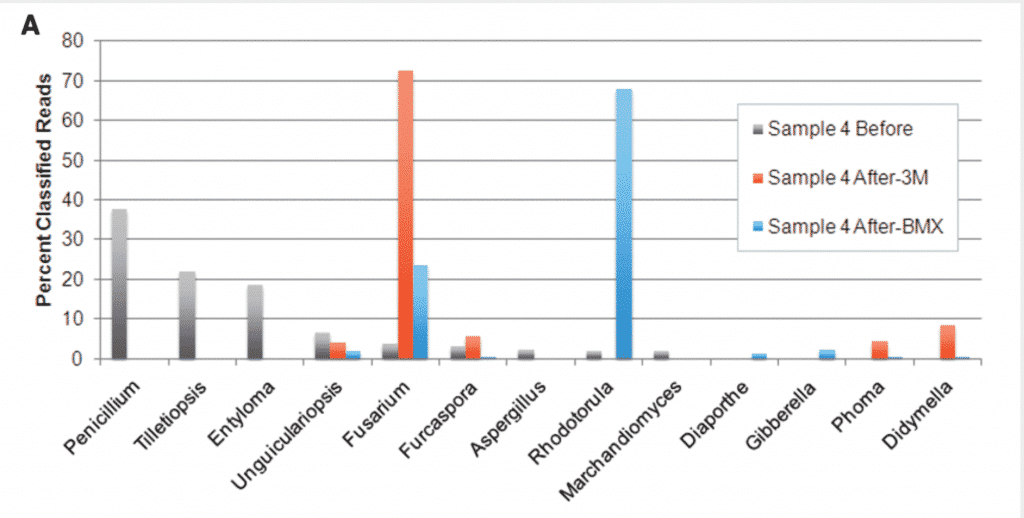
Reproduced from McKernan et al.
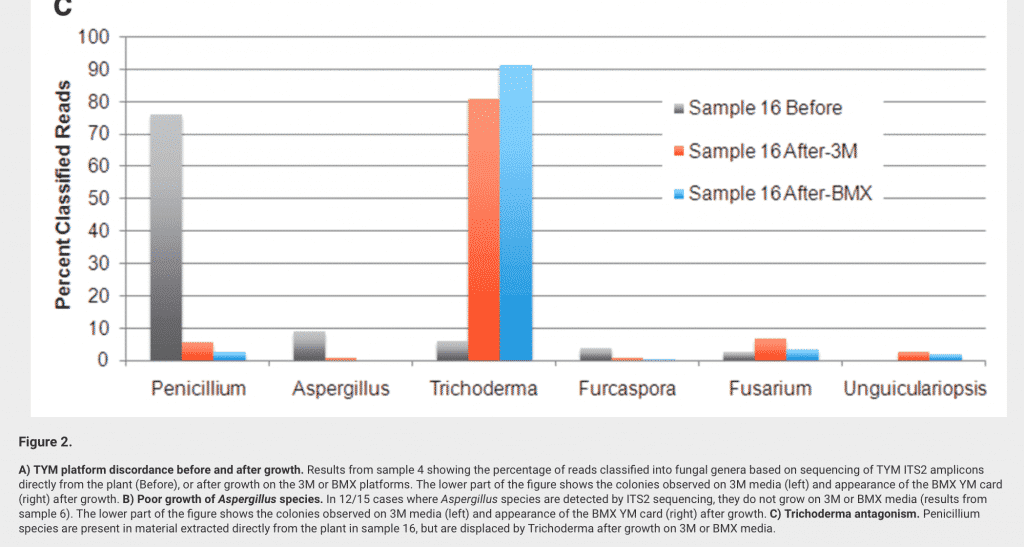
Sample before are sampled directly from the Flower (Grey). After culture, other species dominate.
A more likely reason for lab to lab discordance testing for Aspergillus is sampling bias. Since filamentous fungi severely clump in their growth phase, its very easy to subsample a culture and get all of it or none of it. This is explored on our Heterogeneous MacroColony page.
Another reason for disagreement is likely related to the fact that Aspergillus is a plant EndoPhyte. Many bacteria of concern are also hemp endophytes. Endophytes live inside the plant and most homogenization protocols used for plating gently soak the plant in broth. The gentle soaking is advised to maximize “viability” of the cells coming off of the plant. These gentle homogenization methods cannot lyse open the plant cell walls without lysing the bacterial and fungi cell membranes.
As a result, most plating methods cannot adequately survey the endophytes of the plant. DNA based methods lyse all of the cells open and measure endophytes and epiphytes simultaneously. Since most cannabis flower is ground into a fine powder before smoking the endophytic fraction is an important risk factor to consider.
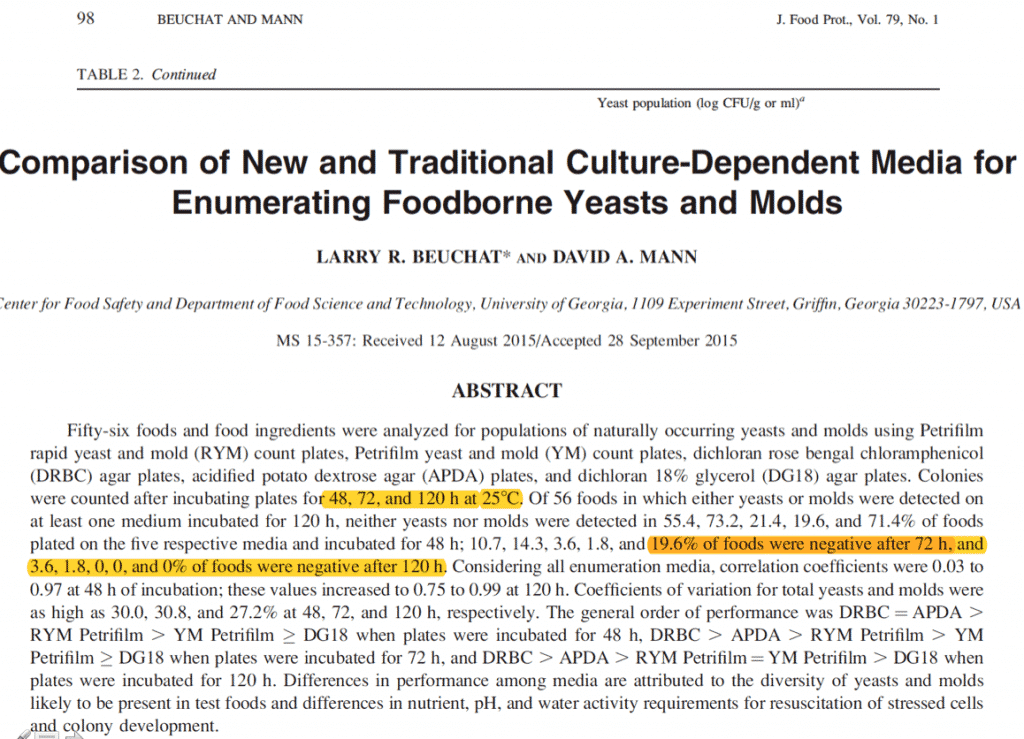
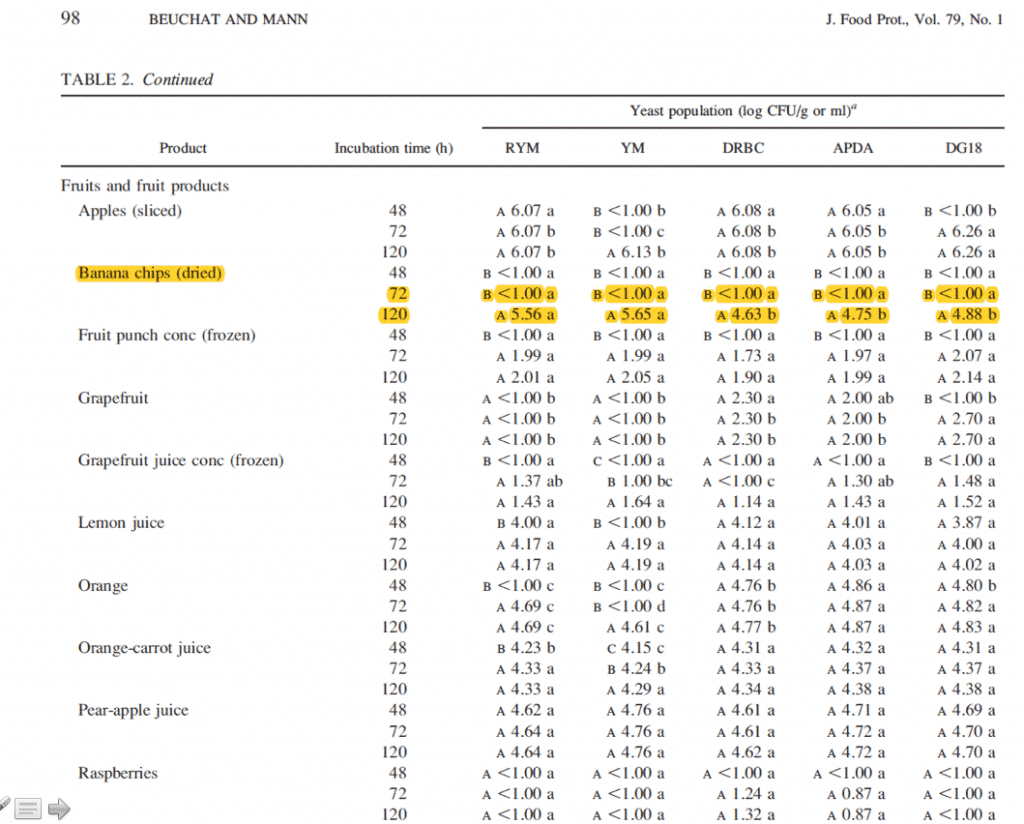
The pursuit of “viability” in a 48 hour culture based test is a mirage. Published literature demonstrates the need to grow for 120 hours to fully resuscitate spores from dried foods (tested across 5 medias) thus the 48 hour testing cannot adequately assess viability and this false viability pursuit sacrifices endophytic detection of Aspergillus. It is important to note that Cannabis is dried and should be treated as a dried food yet no one we know of in the cannabis testing field is running plates for 120 hours. We also suspect attempts to implement 120 hour growth, will only leave one questioning what remains on the sample being testing 120 hours later. Did the original microbiome change in that 5 day testing period?
In summary, there are a host of reasons one can get discordant results testing for Aspergillus. Visual mis-identification is a major source of discrepancies in the field. The first thing to do to troubleshoot this is to rule out sampling bias. Then look for another DNA based method to confirm the results. Plating based methods will deliver biased culturing of the microbiome and will fail to sample the endophytic microbiome. These culture based tools are unlikely to resolve the difference but may be helpful to isolate a species for whole genome sequencing.
Beuchat et al describe the challenges in resuscitating dried spores.