In our most recent publication we demonstrate that Aspergillus appears to have poor viability on various culture mediums thus having 100-1000 fold under-estimate of CFUs on TYM petrifilms compared to qPCR. This is likely due to Aspergillus forming heterogeneous macro-colonies in solution and thus single CFUs are actually derived from a clump of 100s to 1000s viable cells as show by de Bekker et al.
McKernan et al. Figure 3.
Below we have plated Aspergillus stocks in solution and then performed serial dilutions on SabDex Agar plates and rapid TYM petri-films. The stock solution generates qPCR signal that implied 1M CFU/gram and was diluted 1:10, 1:100, 1:1000. These correspond to 23.3 Cq ,26.6Cq, and 29.9 Cq. 10,000 CFU/g doesn’t show up on Agar and has 3-4 colonies on petrifilms. This is a result of heterogeneous macro colony formation in solution prior to seeding the Agar lawns and as a result the CFU counts for Aspergillus are orders of magnitude less sensitive than PCR.
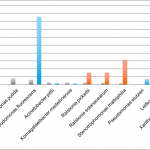
Taking these cultures of Aspergillus and placing them on a 40X microscope and hemocytometer after dilution into ddH20 before plating and one can see mycelium networks and heterogenous macro-colonies. These clustering of cells prior to plating destroy the quantitative accuracy of measuring CFU/g with culture based methods.
Examples of Heterogenous Macro Colonies from Bekker et al.
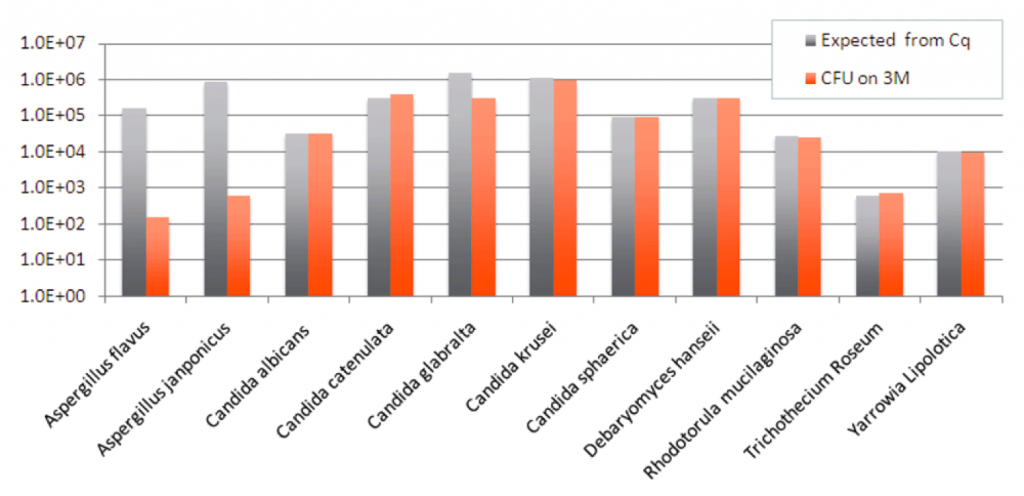
To properly calibrate a qPCR assay for Aspergillus, one must first harvest individual spores and count them on a hemocytometer and perform serial dilutions of those spores into qPCR to measure the response rate of the qPCR assay. Its important to do this with declumped and filtered spores.
Dangerous Aspergillus species cannot be accurately quantitated with culture based methods and should not be used for cannabis safety testing.
©2024 MEDICINAL GENOMICS CORPS
Copyright all right reserved.
Signup to receive email updates on blog posts, podcasts, webinars, new products, and more!
Join us at the CannMed 24 Innovation and Investment Summit, an exclusive experience of discovery, development, and networking with innovators from around the world.