A lab recently posted a study that questions qPCR’s ability to quantitate microbial contamination on cannabis. Although their intentions are good, we believe the authors’ conclusions are based on poor assumptions and not using the best tool available. As a result, the study has created confusion in the marketplace. We’d like to take the opportunity to challenge some of the key points made by the authors.
Fact: The authors did not use the best qPCR tools for the job
First, we are certain the “commercially available” assays the authors used were not Medicinal Genomics kits. We make this conclusion based on the information provided in the supplemental data and the fact that our kits solve for many of the performance issues the authors experienced. More on that below. Had the authors used our SenSATIVAx and PathoSEEK kits, which we specifically designed for the cannabis matrix, we believe their results would have been much better.
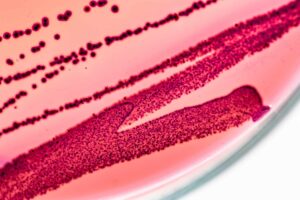
Myth: Plating is the “gold standard” for microbial testing
The authors conclude that because qPCR results do not always match plating they must be incorrect. For instance, they say plating monocultures of bacillus and pseudomonas gives a different CFU value than qPCR predicts. But the problem is that “plating” is a broad term that can include a variety of media types. And those different plating types often don’t agree with each other because microbes replicate at different rates based on the medium they’re grown in (see image below).
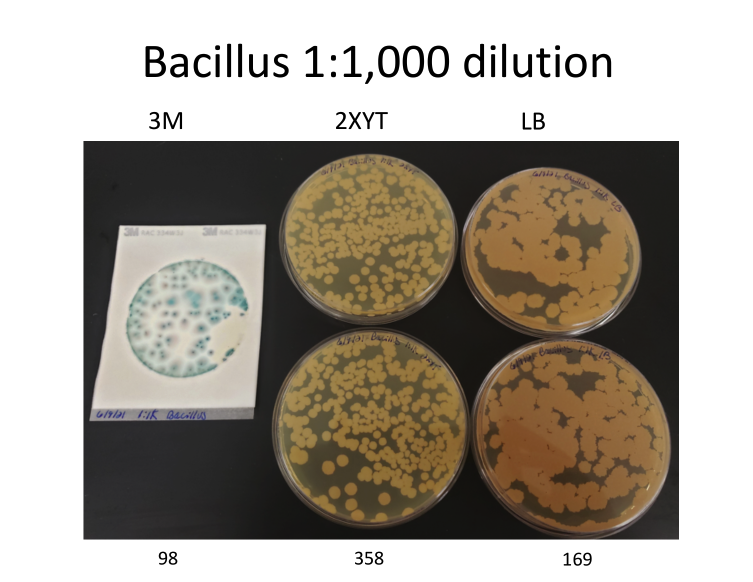
Myth: Variation in ITS copy numbers presents a problem for qPCR quantification
Yes, it is true that our Total Yeast and Mold and Total Aerobic Count assays target ITS regions of the genome. And yes, some organisms have more than one copy of that region. However, plating has a similar problem because not all microbial species replicate at the same rate. Furthermore, plates grow off-target organisms that can inflate their CFU results.
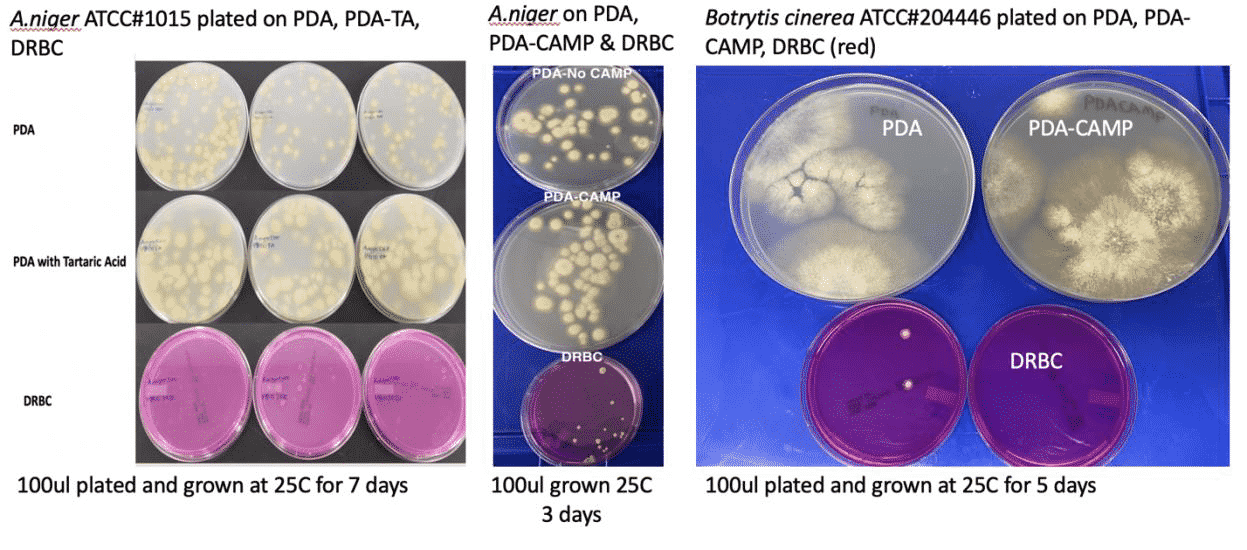
Myth: “Commercial available” assays are contaminated
The “commercially available” qPCR assays the authors used in the study produced strong signal in the no template controls. Based on this finding, we are sure the authors did not use our PathoSEEK assays. Our reagents are NTC tested and certified as clean. The contamination the authors saw was likely due to poor primer design. Our investigation into the primer sequences the authors made public showed a number of issues. Most notably is the fact that the primers hit the cannabis genome. Again, that is not a problem you will find in qPCR assays that were specifically designed for cannabis.
Fact: Cannabis compounds inhibit qPCR
This is true, which is why we recommend labs use our SenSATIVAx DNA Purification Kits to wash away all the compounds and extracellular material found in a sample, leaving behind purified DNA. Other qPCR methods that use a simple boil prep often run into inhibition issues. Furthermore, plating techniques have no purification step to deal with the antibiotics and citrates that are native to cannabis and can throw off their results.
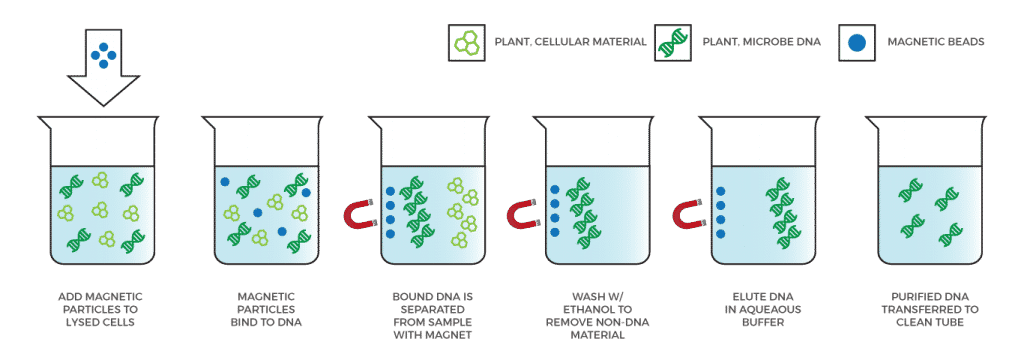
Myth: DNA from dead organisms inflates qPCR results
Many critics falsely assume DNA-based methods will fail more material than culture-based methods when it comes to threshold tests, and leave growers hopeless to employ sterilization techniques. However, scientists, including the team at Medicinal Genomics, have developed techniques to solve the live-dead problem.
Join Our Webinar to Learn More
We plan to address each of these issues and more in our June 30th webinar “Quantitative Enumeration of Yeast and Molds: Myth vs Fact”.