One of the common objections to using qPCR for microbial testing is the fact that the method does not distinguish between live and dead DNA. PCR primers and probes will amplify any DNA in the sample that matches the target sequence, regardless of viability. This is what we call the Live-Dead problem.
Many critics falsely assume DNA-based methods will fail more material than culture-based methods when it comes to quantitative tests and leave growers hopeless to employ sterilization techniques. After all, sterilization kills the microbes but doesn’t eliminate the DNA, so dead organisms will inflate results. However, scientists, including the team at Medicinal Genomics, have developed techniques to solve the live-dead problem. Generally, these solutions fall into two categories: live-dead qPCR and before-after culture qPCR.
Live-Dead qPCR
Live-dead qPCR methods use reagents or dyes to eliminate free DNA or make it so the PCR primers can’t amplify it.
Grim Reefer®
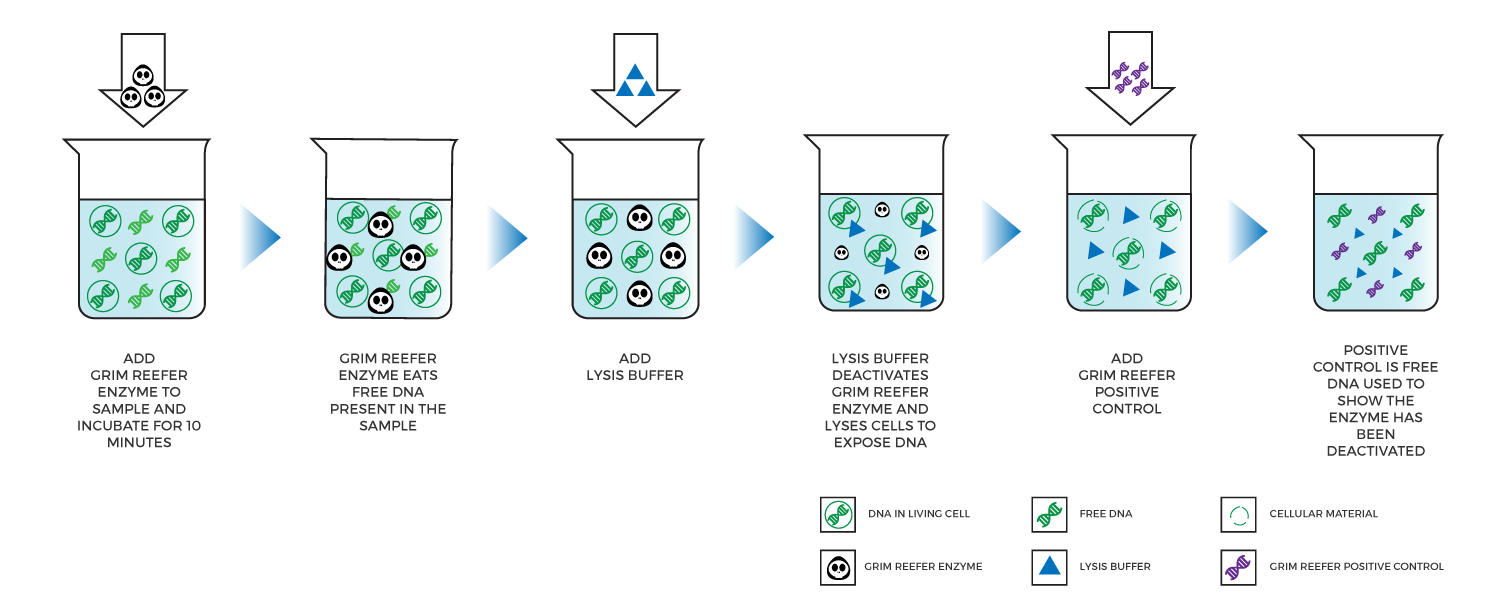
Medicinal Genomics developed the Grim Reefer® Free DNA removal kit, an enzyme-based solution that eliminates free DNA contained in a sample by simply adding an enzyme and buffer and incubating for 10 minutes. Next, lysis buffer is added, which instantaneously inactivates the Grim Reefer® Enzyme so it does not continue to eliminate DNA when viable cells are lysed.
Labs can use Grim Reefer® to treat a sample that fails a threshold test. As a result, Grim Reefer® only needs to be performed on 10-20% of the samples tested. If the results of the Grim Reefer® treated sample are similar to the original, that means the organisms were viable. If the results shift significantly, then DNA from dead microbes likely inflated the original result.
Other Live-Dead Enzymes
There are other enzyme-based free DNA removal kits on the market. However, users must heat the sample at 95C for 15 minutes to inactivate the enzyme. At 95C, live cells will also die, exposing their DNA. That means the enzyme can consume DNA from lysed, viable cells before it is inactivated.
Intercalating Dyes
Other methods use intercalating dyes (or membrane impermeable dyes) that crosslink DNA, making it so PCR primers cannot amplify it. These dyes cannot traverse the cell membrane of a viable microbe, so they only crosslink the free DNA from dead organisms. As a result, dead DNA cannot contribute to a qPCR signal. PMA is a common dye used for this and it requires UV light to cross link the DNA.
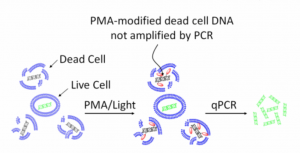
The intercalating dye’s ability to traverse the cell wall is very organism dependent. As a result, there has yet to be a paper demonstrating how to perform this technique on an entire genera like “Total Yeast and Mold”. Most of the examples we’ve found in the literature appear to be highly optimized for a single organism. Furthermore, dyes are expensive, usually teratogens, and hazardous. This does not fit a green chemistry philosophy.
Method 2: Before-After incubation qPCR
Another simple solution to the live-dead problem is to perform a second qPCR run after a short incubation. If the microbes are living, they should replicate their genomes in 20 minutes to 4 hours. Therefore, qPCR results post incubation would show growth. The PathoSEEK Total Yeast and Mold and Total Aerobic Count assays target the ITS regions of the genome, which are high-copy regions. Therefore, the shift in signal is easy to detect even if the cells are partially replicating. This method is often referred to as Before-After incubation qPCR.
Most labs that employ this technique, only perform the 2nd PCR on failed samples. Since labs can perform qPCR in just a few hours, they can get a result before incubation is complete. As a result, after incubation qPCR only needs to be performed on 10-20% of the samples tested.
There are many published methods on Live-Dead PCR. In fact, the EPA recommends qPCR with and without a few hours of growth to capture Live-Dead PCR results. Furthermore, some labs increase the sensitivity of this approach by targeting the ITS preRNA sequences, which are even higher-copy regions.
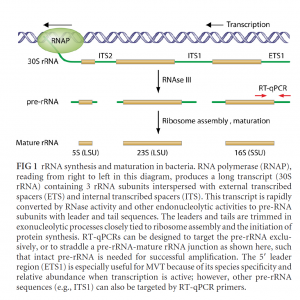
Cangelosi et al. AEM
Sterilization Does not Equal Safety
Although solutions to remove non-viable DNA help labs get a better picture of what is living on the sample, not all microbes are risk due to their viability. Certain species of Aspergillus are a risk because they synthesize pasteurization resistant spores that can lead to a debilitating invasive infection, called aspergillosis.
If you want to HIDE your risks, you may sterilize cannabis so it passes a microbial test, but that masks the very risks labs are trying to measure. That’s why we support regulations that require species-specific testing for pathogenic microbes, such as Aspergillus (A.flavus, A. fumigatus, A. niger, and A. terreus), in addition to total count tests.