The Boston Globe recently highlighted an important scientific debate ongoing in the Cannabis testing market of Massachusetts. Daniel Adams wrote this excellent piece on August 21st. On September 14th, 2018 Analytical Cannabis published an article echoing many of these same sentiments. The debate is centered on the validity of using qPCR versus culture-based plating for Cannabis microbial testing in Massachusetts. The state has approved two labs, one using qPCR and one using culture-based plating, which has “ignited debate over whether the current system is open to abuse by producers who can ‘shop around’ for favorable results”.
As industry leaders in cannabis genetics and pathogen safety, our scientists at Medicinal Genomics strongly believe that DNA-based microbial testing is superior to culture-based plating. Science has proven that traditional methods are unable to detect the vast majority (roughly 98%) of microbes. As a result many cannabis pathogens do not form colonies. Yet current Massachusetts regulations fail to identify this important microbial distinction, causing many pathogens to go undetected when using traditional and obsolete plating methods.
Medicinal Genomics feels this is a significant liability for the cannabis industry as a whole. As a result, major testing labs are not properly detecting these common pathogens, exposing patients and cannabis employees to undue health concerns.
Our position on this issue is expanded in our recent letter to the Massachusetts Cannabis Control Commission, requesting regulation adaptations to reflect more accurate and more specific DNA-based techniques that can detect any and all common microbes. In addition, we’ve written a substantial review of this topic in a document prepared for the United States Pharmacopeia.
When articles include statements that are directly against what we know to be scientifically proven, we feel obligated to clarify the facts. There are a number of statements made in the Analytical Cannabis article that are not 100% scientifically accurate. The statements stem from Chris Hudalla, a well-respected scientist in the cannabis testing market. We have worked closely with Chris over the years, and ironically, co-authored a manuscript on this very topic with him. MGC has great respect for his contributions to the field, but we feel his comments may have been taken out of context. We feel a comprehensive rebuttal is the most transparent approach to such a complicated topic.
NOTE: The reader should be aware that we have a financial interest in this story as we manufacture one of the technologies being discussed (qPCR).
Within this blog post, we have identified the statements included in the Analytical Cannabis article that Medicinal Genomics knows to be false, those that are true, as well as supplemental data on our position on each specific issue.
- False: The FDA and Other Industries Only Endorse Culture-Based Plating
- False: Food Testing Techniques are Directly Applicable to Cannabis
- Correct: Plating Methods Grow Off-Target Organisms
- False: Testing Results Must be Reported in CFU/g
- Summary
False: The FDA and Other Industries Only Endorse Culture-Based Plating
At one point in this article, Chris Hudalla makes the assertion that (only?) culture-based plating has the backing of the cosmetic industry and the FDA. When he says, “The method they are using is accepted by nobody” he is referring to qPCR.
Let us address the first argument Chris has presented, which is that other industries such as cosmetics, and the FDA endorse one technology.
The problem with this appeal to antiquity (how it has always been done) is that cannabis has been illegal since 1937 and this prohibition predates the existence of the FDA. “How it has always been done in food” is misleading logic as it has never been done before in Cannabis and Cannabis is not food.
The food and cosmetic industries use qPCR for many clinically regulated tests. Below, we have provided a list of references to qPCR being used for FDA approved tests. In addition to many qPCR based tests being approved by the FDA, 98% of the clinical tests being run today are non-FDA approved Laboratory Developed Tests (LDTs). These tests make life and death decisions for human health care in the CLIA lab settings and they have remained outside of FDA review to ensure rapid innovation. More importantly, FDA approval is not required for these LDT tests nor is it required for cannabis testing today.
This appeal to authority actually works against the culture-based plating argument as those very authorities have large programs that are funded to get rid of Plating. This field is known as Culture Independent Diagnostic Testing or (CIDT).
Both the CDC and FDA have multiple independent programs to eliminate culture-based plating from the food testing industry due to its failure to accurately assess foodborne illness risk. The FDA program is known as the Genome Trakr network and the CDCs program is known as Pulsenet. Both programs use genome sequencing to survey food outbreaks across a network of labs around the country. This is being done because culture-based plating takes too long and fails to detect too many risks.
In general, agencies are technology agnostic but validation centric. It is important to understand what it means to be validated.
False: Food Testing Techniques are Directly Applicable to Cannabis
Independent of what testing techniques these agencies recommend for food, we cannot pretend cannabis is food. Cannabis is ground up and inhaled, often by immunocompromised patients. Your food is not. Route of administration is critical in establishing tolerable microbial load. The superimposition of the methods that have been validated in the food industry onto a new matrix like cannabis is a major error, and it misleads the public and political organizations.
The second error in the complaint is the assumption that validation work done for culture-based plating in other industries can be superimposed on cannabis. To date, there are no published manuscripts to support using culture-based plating methods on Cannabis. This is an assumption that works well in short-form media but fails under any prolonged scientific scrutiny.
As anyone versed in cannabis testing can assure you, if you change the matrix, you must revalidate your technology. One cannot just superimpose the testing techniques from tomatoes onto Cannabis and assume the method’s function. One must revalidate their methods using a given technology on the new matrix. There are no publications from Chris Hudalla validating his methods on cannabis yet there are two manuscripts published by our group comparing qPCR to various plating methods.
Both of these peer-reviewed studies point to massive clinical liabilities present with culture-based plating technologies. Chris Hudalla is even a collaborating author on one of these papers, and the paper made it very clear that culture-based plating methods are failing to detect real clinical risk while also suffering from very high false positive rates. Chris is correct to highlight that there are discordant results between qPCR and culture-based plating but is incorrect to assume culture-based plating is the gold standard in Cannabis.
There are two reasons these Plating methods fail in cannabis.
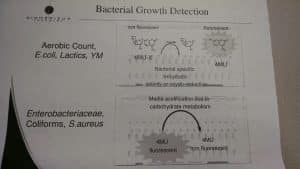
- Plating methods can’t detect endophytes. It is now well published that many of the harmful cannabis pathogens (Fusarium, Aspergillus, etc.) are endophytes. These live inside the plant as shown in this publication (hemp_endophytes). Culture-based techniques need to gently soak the sample in broth so as not to kill any microbes. It is impossible to survey the endophytes inside of cannabis without lysing open cells and culture-based plating cannot detect lysed cells. As a result, culture-based platings’ achillies heal is that it cannot pick up endophytes without killing the organisms on the plant surface (Epiphytes). qPCR can see both.
- Most organisms do not culture and fewer culture in 48 hours. Many fungi need at least 120 hours for spores to culture from dried produce. This has been published on 5 different medias. Any culturing of a dried produce for only 48 hours is security theatre and thus the viability it claims to be capable of estimating is grossly flawed.
While Chris Hudalla is an author on a manuscript highlighting these shortcomings, he has not published anything from his lab suggesting he has addressed these shortcomings of his platform. His public statements seem to differ from the work he put his name on in the scientific record.
Correct: Plating Methods Grow Off-Target Organisms

As mentioned earlier, we have demonstrated that culture-based plating methods grow off-target organisms in several publications.
Most recently, we sent a saturated culture of an antibiotic-resistant Pseudomonas aeruginosa from ATCC to the two testing labs in Massachusetts using culture-based plating. We chose Pseudomonas aeruginosa because it is a Bile-Tolerant Gram-Negative (BTGN) bacteria, and the USP has listed it as required for any valid BTGN test. Massachusetts requires BTGN testing for all cannabis samples.
Both of the culture-based plating technologies used in those labs (Biomerieux and Hardy plates) failed to detect the Pseudomonas aeruginosa in their BTGN tests. However, their Total Yeast and Mold test detected the Pseudomonas aeruginosa when it should not have. After all, Pseudomonas aeruginosa is a bacteria.
Although these results are inaccurate, they are not unexpected. We published in a peer-reviewed journal that over over 60% of the DNA from the growths found on their yeast and mold tests came from bacteria.
In order to design TYM qPCR assays to perfectly mimic the flaws of culture-based plating, we have to intentionally remove the microbes we target with qPCR that do not form colonies and make independent assays for these fungi (more cost). This exposes the State of Massachusetts to the lawsuits they are currently seeing with employees being exposed to high level of unculturable but sickening microbes.
If these platforms cannot detect the proper organisms from a saturated culture without any cannabis present, you cannot rely on the results they give you with matrix or cannabis present. These methods lack proper controls and spike in experiments. Likewise, there is not any published validation data for their microbial methods. For public safety, it is imperative labs have transparent methods and preferably peer-reviewed.
Transparency is critical for patient safety
While we do our best to publish data regarding our methods, any laboratory using our reagents is expected to revalidate in their laboratory setting. This would include proper inclusion and exclusion analysis with live ATCC controls so the public knows what the lab can detect and what it cannot detect. These same studies must be performed in the presence of cannabis matrix to understand how this oily matrix (that often contains 30% antimicrobial cannabinoids) alters the growth of various pathogenic threats. This is a pillar of ISO 17025, which is required for Massachusetts testing compliance.
It is very clear to us that the Massachusetts cannabis testing labs using culture-based methods have not performed these spike-in controls or they would have seen the incorrect results. BTGN results from Massachusetts labs using culture-based plating methods have been unreliable for nearly 4 years now. This can negatively impact Cystic Fibrosis patients known to use cannabinoids to manage inflammation.
Furthermore, there are employee lawsuits in Massachusetts for exposure to Powdery Mildew and Botrytis (Yeast and Molds). These do not culture in plating methods, and growers who test with labs using culture-based plating didn’t believe the employees’ exposure. Perhaps if there were public methods and validation data for using culture-based plating on cannabis, we could have avoided these blind spots and employee/employer conflicts.
This is why each lab must perform their own validations and not rely on the manufacturer’s validation work. However, in this case, culture-based manufacturers have published no public data to support the use of their products in cannabis. To the contrary, there are peer-reviewed papers warning of the platforms’ failure to perform with certain spices (tempo_interferance).
False: Testing Results Must be Reported in CFU/g
A final critique that has been voiced in other forums is that the Massachusetts regulations reference the AHP, which uses the metric of CFU/g. Since qPCR doesn’t report in CFU/g, opponents say the method can’t be used.
Insisting qPCR results match a method that fails to perform in certain matrices presents a real and immediate risk to patients and employees.
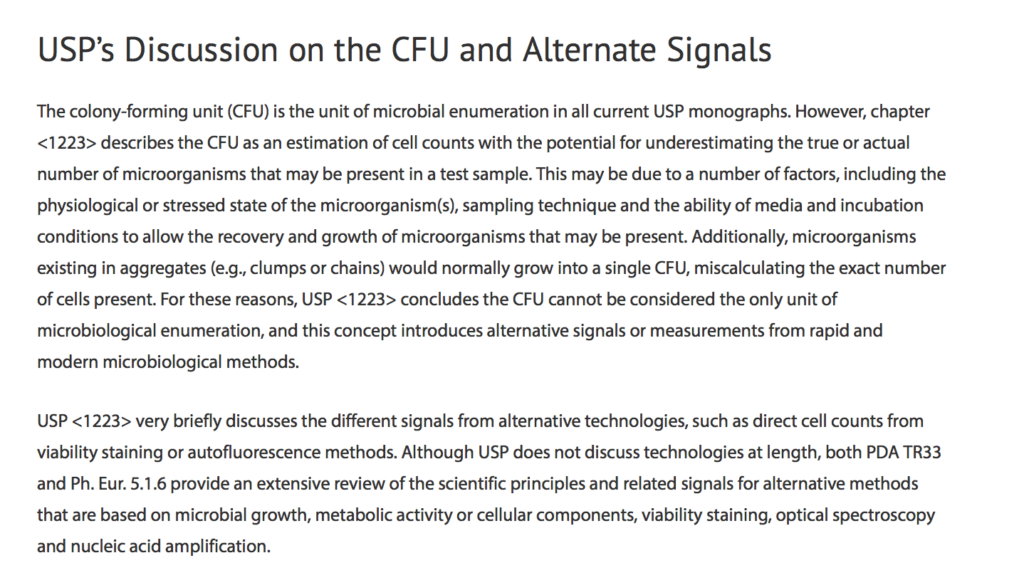
Second, the logic is not supported by the USP, who in their informational chapter <1223> Validation of Alternative Microbiological Methods concluded that CFU/g cannot be considered the only unit of microbiological enumeration.
Lastly, one of the commonly used culture-based plating platforms in Massachusetts (Biomerieux) doesn’t form colonies either. It measures a fluorescent change based on a pH shift.
qPCR also uses a fluorescent signal, but the signal is based on far more specific DNA signatures than a simple pH shift. Many acid producing bacteria can falsely alter a pH-based readout. We demonstrated this with Pseudomonas aeruginosa.
This near-religious attachment to CFU is not reflected in other regulatory agencies that allow other technologies to provide equivalent metrics. This insistence on a CFU/g metric exposes patients to harmful microorganisms that do not form colonies. This has been peer-reviewed and published. Many of the Biomerieux shortcomings are revealed in the document we submitted to the USP.
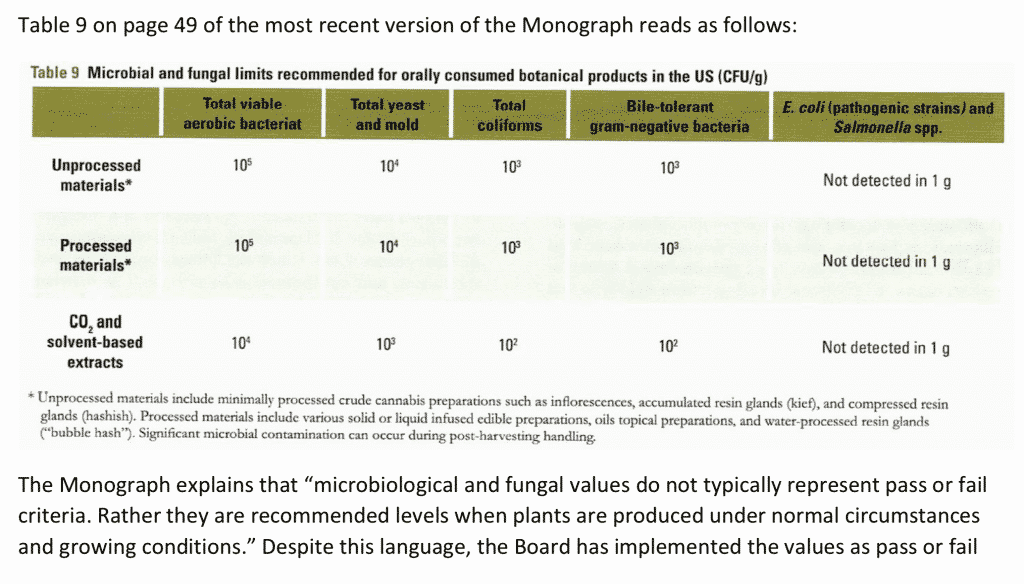
We agree with Chris Hudalla that the regulations as written overly speak in terms of CFU/g. This should change to reflect the USP <1223> language regarding other equivalent or validated technologies. The EP has similar guidance on this. Doing so would enable technologies that can detect the harmful microorganisms that do not form colonies but are creating lawsuits in Massachusetts. This same complaint has been raised in other states struggling to implement the AHP monographs vague definitions. The state of Washington Cannabis Laboratory Association has petitioned for changes to these regulations due to the lack of specificity of CFU/g and the explicit statement in the monograph that these values do not represent pass-fail criteria but are reflective of average microbial loads. Alaska and California have abandoned these requirements and moved to species-specific, presence or absence regulations.
Combine the AHPs lack of trust in CFU/g being an adequate pass-fail criteria and that of the USP <1223> describing why CFU/g cannot be the only form of microbial enumeration, and the assertion that results must be reported in CFU/g falls apart.
Summary
Medicinal Genomics stands behind the data supporting our scientists’ positions against the identified statements in the September 14th Analytical Cannabis article. As leaders in the ever-changing world of cannabis – safety, science, and education are three areas of focus that have to be right. The viability and integrity of our company depend on it.
We understand the evolution of cannabis science and its proper implementation will take time. Labs, growers, and regulators are venturing into unchartered territories. Our goal at Medicinal Genomics is to provide accurate scientific data to the field, allowing cannabis professionals to make informed decisions that will keep patients and industry employees safe. Through our transparent scientific approach, we have demonstrated that DNA-based techniques are the best detection defense against unculturable pathogenic microbes – a position also supported by the FDA and the CDC. No other microbial platform manufacturer has published a more thorough evaluation of their method on cannabis as we have in our validation document (download here).
Attempts to undermine qPCR’s abilities to detect microbial contamination on cannabis are not only unfounded, but they also put patient safety at risk. Culture-based plating methods are antiquated and unable to detect the vast majority (98%) of microbes that do not form colonies, and we feel this is a danger not only to consumers, but to the cannabis industry at large.
There are two labs in Massachusetts (CDx Labs and EVIO Labs) that have received ISO17025 accreditation with our qPCR methods.
There are labs in Pennsylvania (Steep Hill Labs) that have received ISO17025 accreditation with our qPCR methods.
There are over 100 labs in the US and Canada that use these qPCR methods.
Our Validation documentation is public on our website here.
We have submitted much our work to the USP expert review panel on this topic.
Peer Reviewed manuscripts for our qPCR being used on Cannabis.
Authors other publications in microbiology
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC187521/
- https://www.ncbi.nlm.nih.gov/pubmed/18775913
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC373392/
qPCR Methods Used in Other Industries
Food Methods
- Diagnostic Real-Time PCR for Detection of Salmonella in Food: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC535175
- Rapid detection of Salmonella in foods using real-time PCR. https://www.ncbi.nlm.nih.gov/m/pubmed/19256088/
- A quantitative PCR assay for the detection and quantification of Shiga toxin-producing Escherichia coli (STEC) in minced beef and dairy products. : https://www.ncbi.nlm.nih.gov/m/pubmed/21878400/
- Rapid, Specific, and Sensitive Detection of Spoilage Molds in
Orange Juice Using a Real-Time Taqman PCR Assay: http://jfoodprotection.org/doi/abs/10.4315/0362-028X-69.2.385 - foodProof Yeast & Mold Quantification in Dairy Products: https://www.bc-diagnostics.com/products/kits/real-time-pcr/spoilage-organisms/foodproof-yeast-mold-quantification-lyokit/
AOAC Validated qPCR Methods
- Bio-Rad’s iQ-Check Salmonella II Real-Time PCR Kit Earns Approval as an AOAC Official Method of Analysis: http://www.bio-rad.com/en-us/food-science/news/bio-rads-iq-check-salmonella-ii-real-time-pcr-kit-earns-approval-as-aoac-official-method-analysis?ID=Bio-Rad-s-iQ-Check-S_1519852048
- AOAC Certifies BAX® System Real-Time PCR Assays for Non-O157:H7 STEC in 25g Flour and Ground Beef: https://www.hygiena.com/hygiena-receives-aoac-certification-for-bax-system-real-time-pcr-assays-for-detecting-of-non-o157-h7-stec-in-25g-flour-and-ground-beef.html
Clinical Microbiology
- Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1360278
- A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5288344/
- Real-time PCR as a diagnostic tool for bacterial diseases: https://www.ncbi.nlm.nih.gov/m/pubmed/23153240/
- Quantification of Intestinal Bacterial Populations by Real-Time PCR with a Universal Primer Set and Minor Groove Binder Probes: a Global Approach to the Enteric Flora: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC427818/
- Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set: https://www.ncbi.nlm.nih.gov/m/pubmed/11782518/