In 2020, a Michigan lab contacted us claiming our CFU/cq calculations were not matching their samples, nor did they match a recently released Candida albicans NSI Total Yeast and Mold (TYM) standard.
First, it should be noted that there are a number of reasons why qPCR results and plating results may disagree when the methods are compared in a real-life scenario, including:
- Homogenization and sampling technique
- Endophytes
- Off-target growth
- Regional differences in microbiomes
- Insufficient Standards
You can read more about those items in our Knowledge Base Article titled, “Why do PathoSEEK® Results Sometimes Disagree with Plating Results?”.
To better understand these discrepancies, our lab agreed to investigate. Due to federal law, the lab could not share flower samples across state lines, so they shipped Petrifilms to our lab. We selected 20 representative colonies from those plates and extracted the DNA for further examination. Of those 20 colonies, 9 produced enough quality DNA to perform whole genome shotgun sequencing on an Illumina HiSeq platform. We analyzed the reads with two different bioinformatics pipelines. SPades was used to assemble the genomes and BLAST against NCBI. OneCodex (a 3rd party independent analysis company) was used to perform an assembly free Kmer classification of the genomes. The results are listed in the below Table. The Whole genome assemblies are hyperlinked as fasta file and the OneCodex reports are hyperlinked as public documents.
| Michigan Sample | Spades Assembly | OneCodex Analysis | |
| 12283-1-5_001.fastq.gz | Bacillus.fasta | Bacillus velezensis | Bacteria |
| 12335-1-3_001.fastq.gz | Staph.fasta | Staphylococcus haemolyticus | Bacteria |
| 12335-1-4-LG_001.fastq.gz | Penicillium.fasta | Penicillium citrinum | Fungus |
| 12339-1-3_001.fastq.gz | Staph.fasta | Staphylococcus epidermidis | Bacteria |
| 12339-1-4_001.fastq.gz | Staph.fasta | Staphylococcus epidermidis | Bacteria |
| 12352-1-1_001.fastq.gz | Bacillus.fasta | Bacillus amyloliquefaciens | Bacteria |
| 12352-1-5_001.fastq.gz | Bacillus.fasta | Bacillus amyloliquefaciens | Bacteria |
| 12352-1-6_001.fastq.gz | Bacillus.fasta | Bacillus amyloliquefaciens | Bacteria |
| 12883-4_miseq.fastq.gz | P.brevicompactum | Penicillium sp. MA 6036 | Fungus |
As you can see, seven of the nine colonies (78%) were Bacteria and not a Yeast or Mold. This agrees with our previous publications on this topic where we found that 60% of the DNA from TYM plates are actually bacteria.
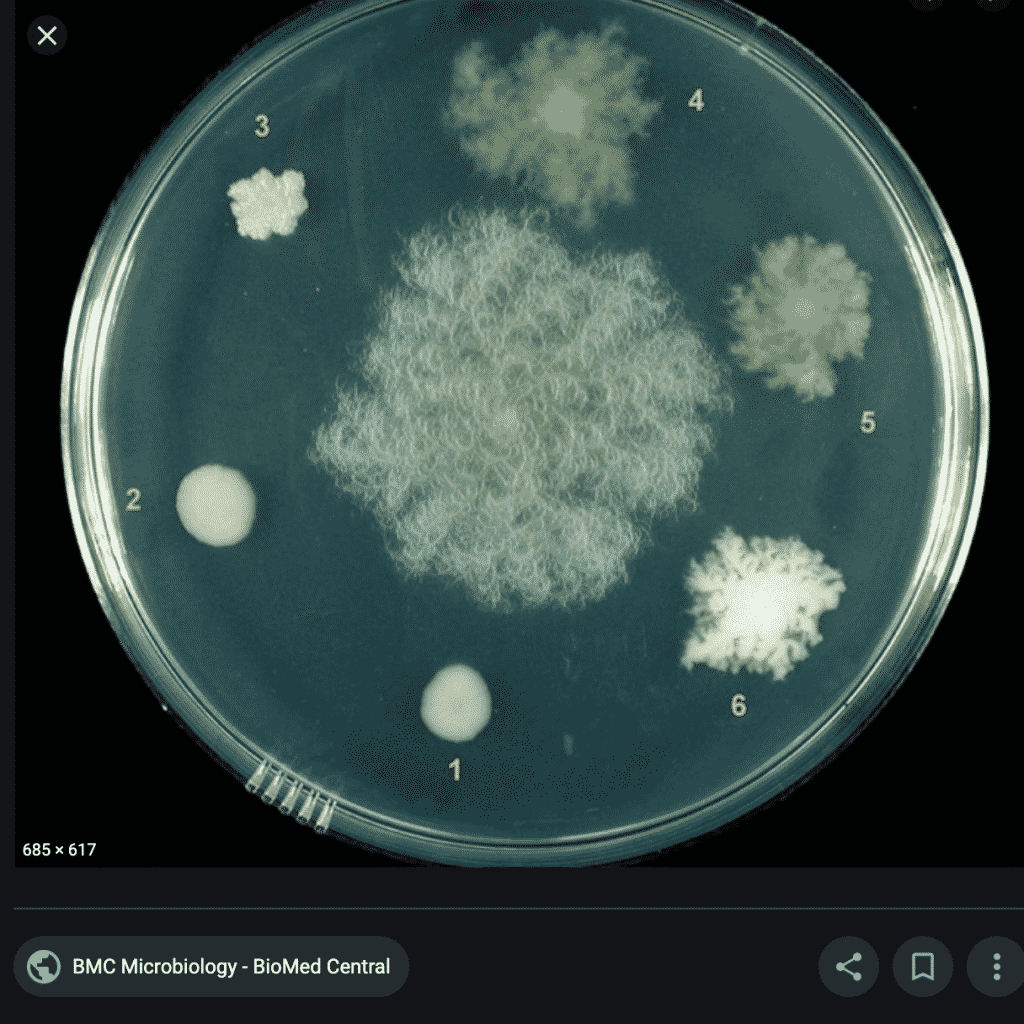
What do these colonies look like? Surely a human eye could discern them and avoid including them in the TYM counts?
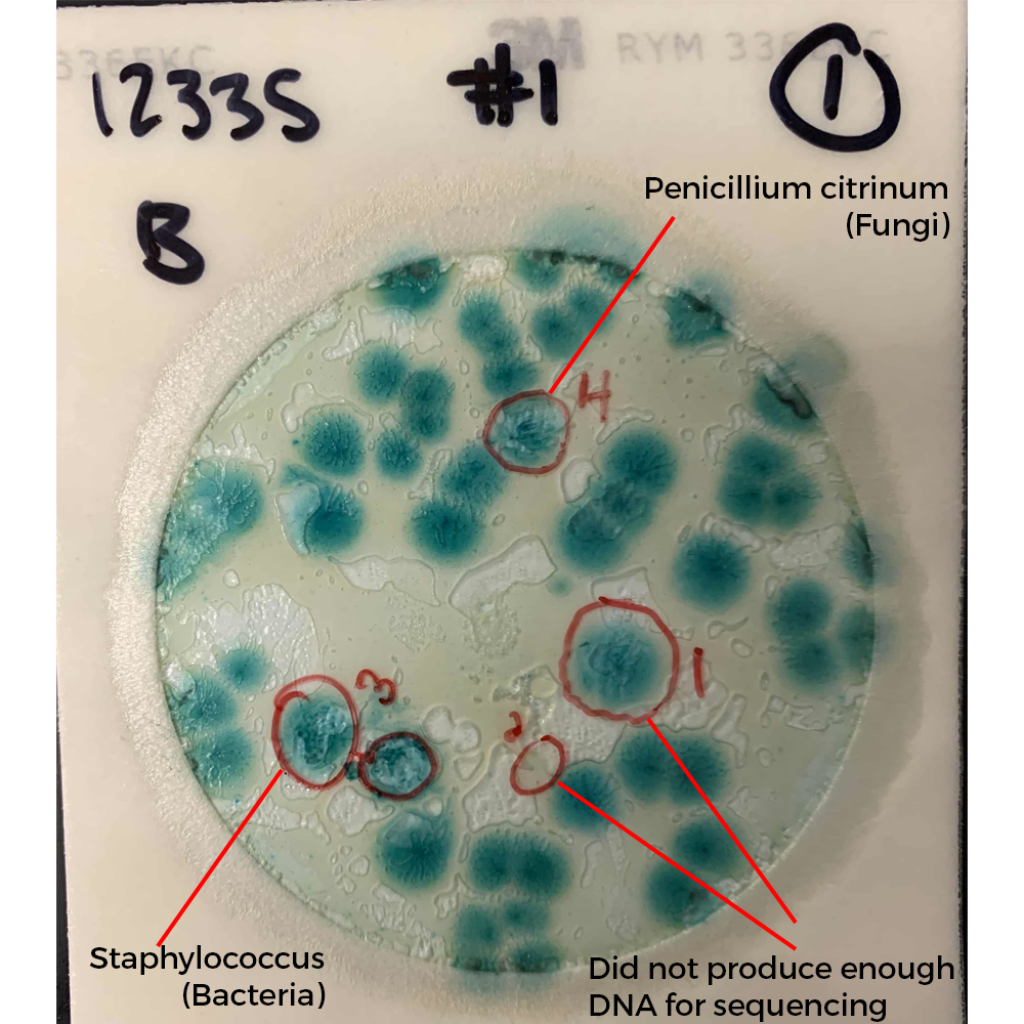
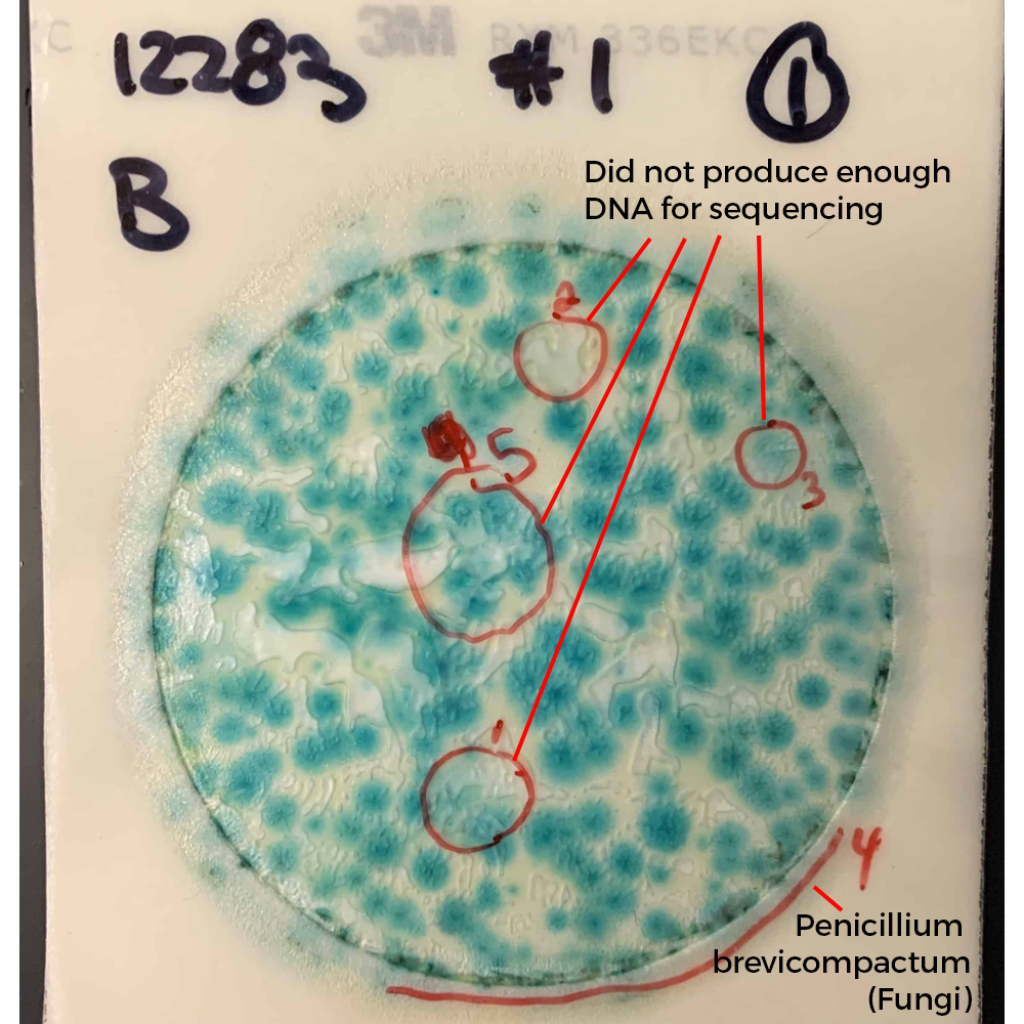
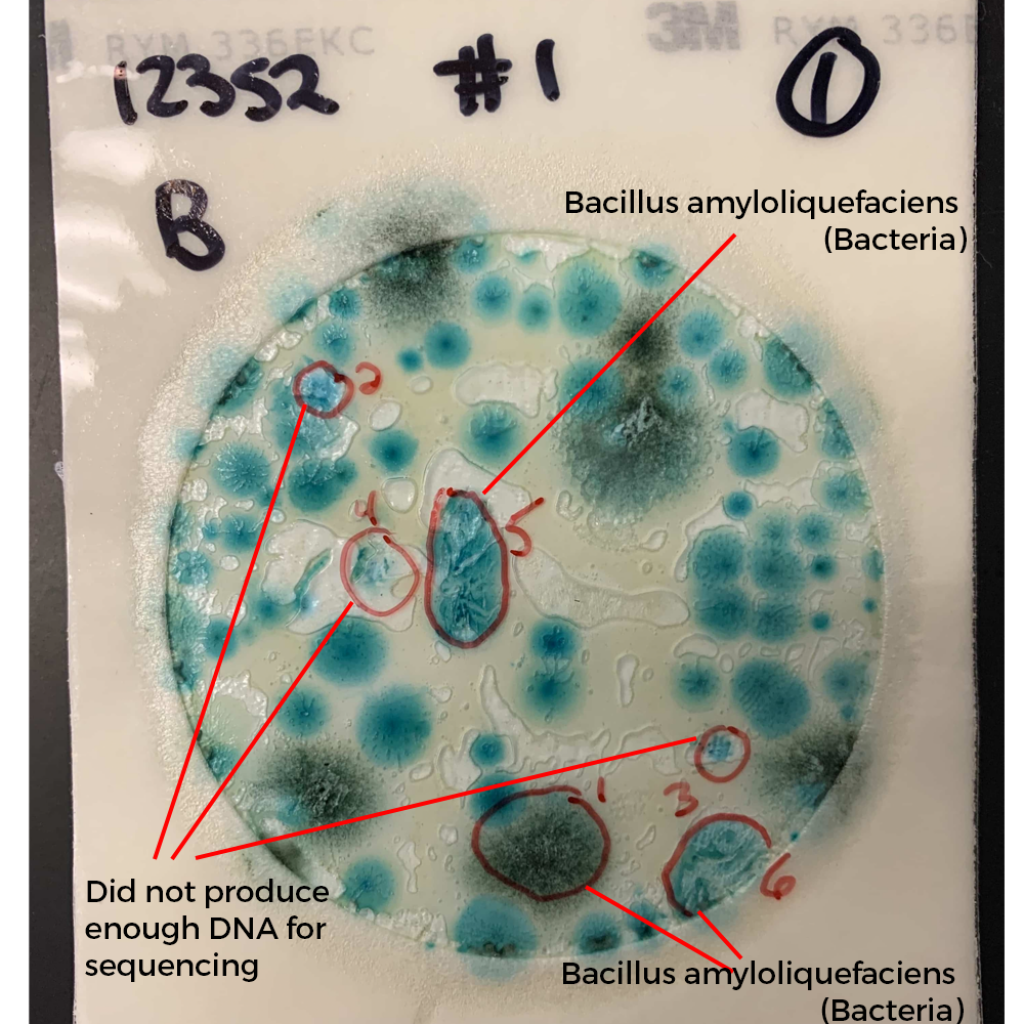
These colonies all appear similar to the eye. A method that doesn’t rely on such subjective interpretation (i.e. qPCR) would be a big help to the industry.
As you can see, any regulation that invokes CFUs as the gold standard metric for safety is a regulation that has no specificity and will result in perpetual lab to lab discordance, arguments, and lawsuits, as no one really understands what it is they are measuring.
This certainly begs the question, what are these TYM plating assays good for? TYM plating assays fail to:
- Distinguish bacteria from fungi
- Discern beneficial microbes from pathogenic microbes.
- Detect 95% of the microbial population that is unculturable
- Detect endophytes that live within plant cells
When gold standards are this bad, we should not be forcing newer technologies to replicate their failed behavior. We should use the right tools with patient safety in mind. We cannot demand the industry use a “gold standard” that is blind to the pathogens that pose the greatest risk, such as Aspergillus. That is an appeal to antiquity not an appeal to patient safety.
Many states are recognizing how difficult it is to ring test labs with TYM assays regardless of the platform each lab utilizes. Even different plating methods provide different results. As a result, many states are migrating to species-specific Aspergillus testing in place of TYM testing. We believe this approach is more grounded in defendable clinical utility.
July 16th Update.
A question was raised regarding the impact of dead DNA on the sequencing analysis. Since Chlorampenicol and Chlortetracycline is used in RYM plates, could there be dead bacterial DNA contaminating the DNA assemblies? This is an excellent question. There are several ways to address this with sequencing data and literature review.
1)There is peer reviewed literature on many of these organisms being resistance to Chlorampenicol and Chlorotetracyclines.
https://jb.asm.org/content/104/3/1095.short
https://aac.asm.org/content/9/6/962.short
https://aac.asm.org/content/35/7/1277.short
Bacillus amyloliquefaciens has been published as an organism that can host the synthesis of chloramphenicol. This would be very difficult to do if the organism was also susceptible to chloramphenicol.
Bacillus amyloliquefaciens has also been published as a bioremediation organism to clean aquatic environments of chlorotetracylclines. This would be a poor choice or remediation organisms if it was also susceptible to chlorotetracyclines. These are the two antibiotics listed on RYM products. While may have B.subtilis listed in their exclusion criteria, these exclusion criteria do not extend to other strains. Even Bacillus subtilis can cary plasmids that confer resistance to an antibiotic in one Bacillus subtilis and not others lacking the plasmids. This is also demonstrated on the two antibiotics listed in RYM products. While the sequencing detected Bacillus amyloliquefaciens, we feel this is an important point to raise with folks who conflate microbial strains due to similar genus names.
As an example, one would not use Bacillus subtilis to run experiments on Bacillus anthraces. The later organism is tightly controlled because its also known as Anthrax. The former is a biocontrol agent used in agriculture. Conflating organisms even on the species level is dangerous let alone the genus level. This is also seen in Cannabis sativa, where both hemp and drug type cannabis are in the same genus and species yet legally and genetically produce vastly different chemotypes.
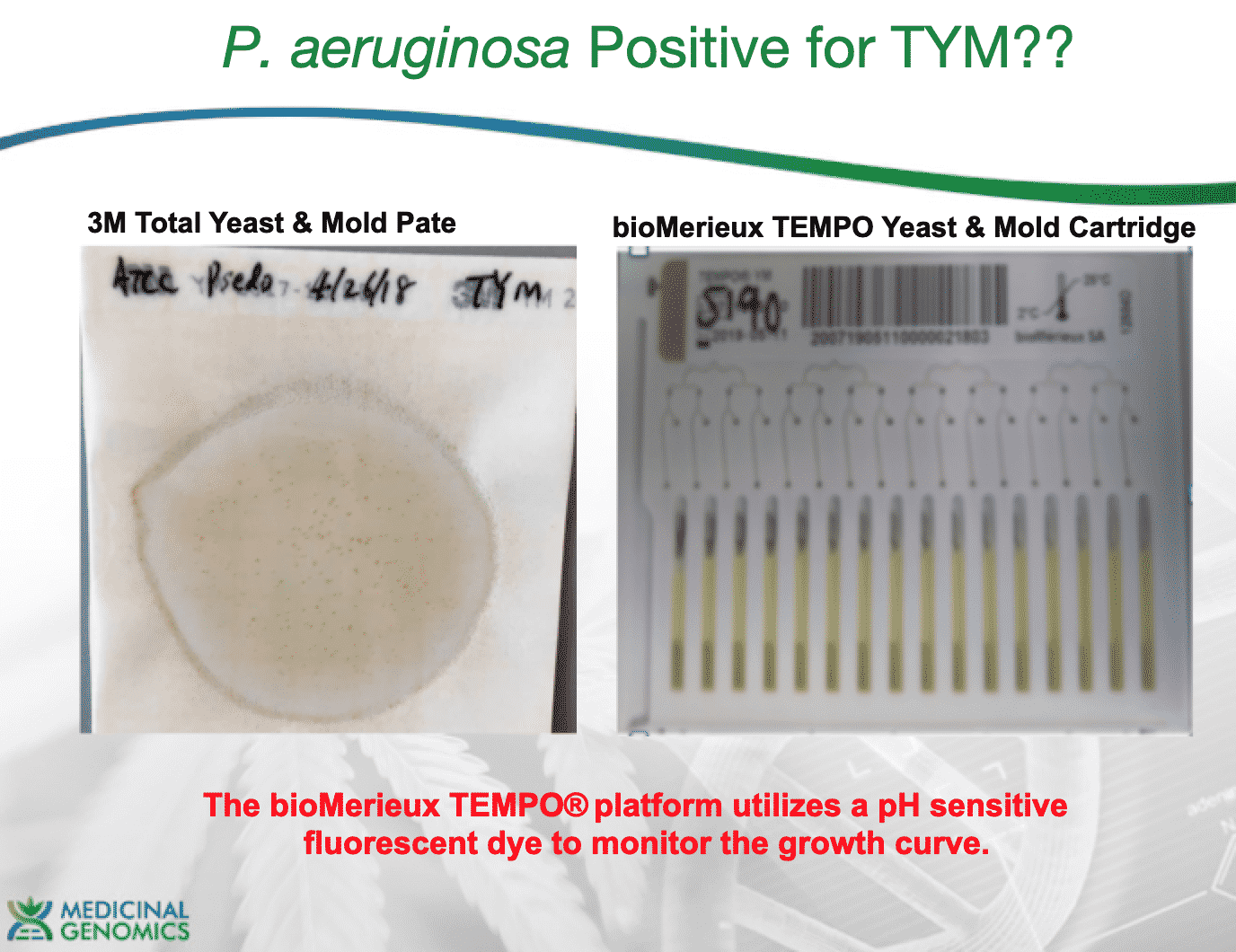
In 2018, we also plated bacteria (Pseudomonas aeruginosa from ATCC) on TYM plates and observed colonies on and false positives on Biomerieux Tempo YM cartridges. P.aeruginosa produces acid and lowers the pH of solutions and can trigger many chromogenic detection systems.
2)OneCodex has a scan for a subset of Antibiotic resistance genes. Every genome except one (the low coverage, large genome (fungi) run on the MiSeq) has at least 1 Antibiotic resistance gene and all of the Staph genomes have 9-12 antibiotic resistant genes including the gene that provides resistance to Chlortetracycline used on TYM.
Secondly, one needs to keep in mind that these antibiotics are not alway specific to bacteria. There is peer reviewed literature showing chloramphenicol inhibiting the growth of Fusarium and Aspergillus. These fungi have documented harm to cannabis patients.
Antibiotics are not a sharp knife and while people are tempted to use them to split bacteria from fungi, they do not have the specificity of DNA primers and can lead to false negatives or unexpected competitive growth environments due to biased selection.
Antibiotic anti-fungal activity:
https://www.sciencedirect.com/science/article/abs/pii/S0168160507003005?via%3Dihub
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2606932/
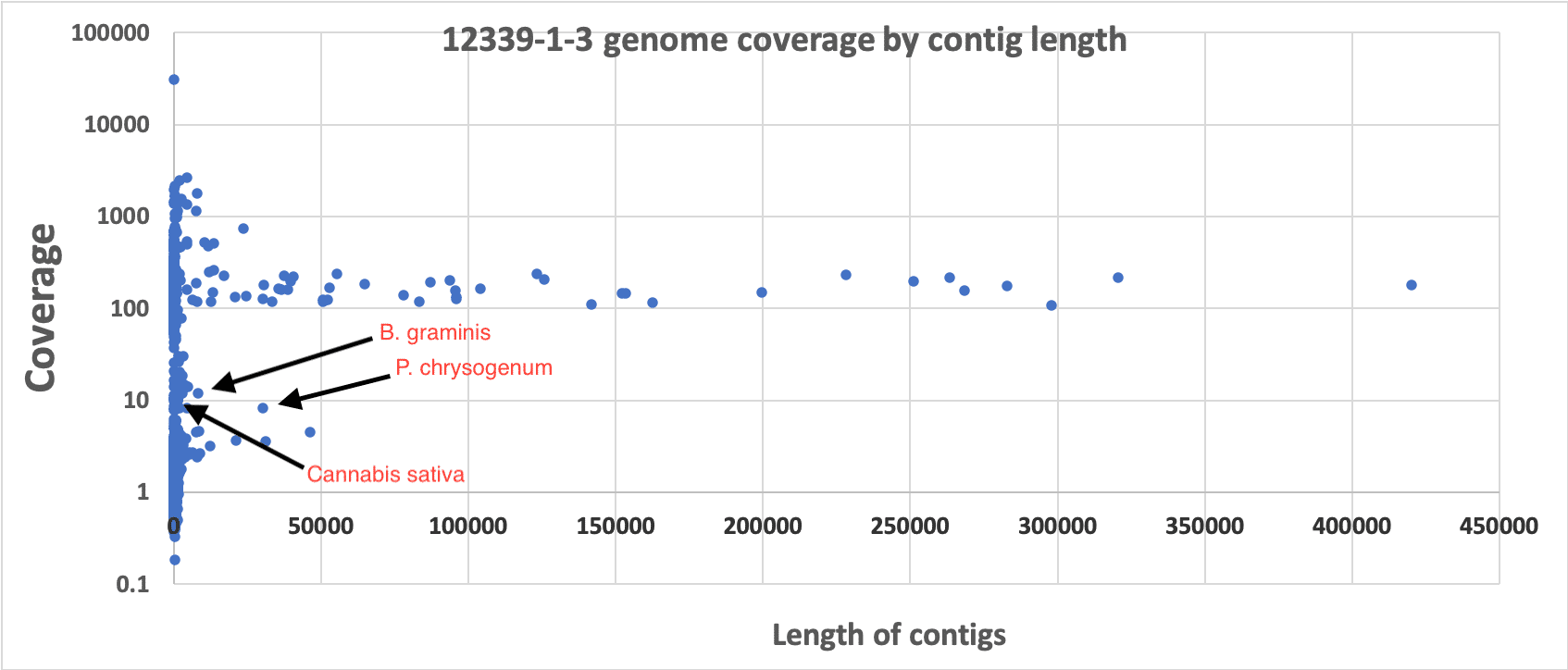
3)Coverage analysis. If dead DNA was a contaminant it would be diffuse and not constrained to a colony like its membrane bound live counterparts. Therefore we would expect it to be low coverage compared to the primary colony that was isolated. We have provided coverage plots by the length of the contigs or chromosomes. These all demonstrate primary coverage peaks at 80-150X coverage comprising the majority of the assembly’s sequence. In some cases, there is a small coverage peak at 4X coverage that might represent metagenomic contamination of dead DNA, low MOI plasmids or endofungal bacteria. BLAST results of these low coverage contigs reveals cannabis DNA. If there was bacterial dead DNA, it would have a similar molarity and copy number of the cannabis DNA and would not form high coverage, highly contiguous primary assemblies.
The other possibility is existence of endofungal bacteria where the bacteria live inside the fungi at high copy number. When we have seen Endofungal bacteria in the past on Cannabis (Ralstonia), its’ coverage is higher than the fungal genome as the copy number of the bacteria in the fungi is higher than the nuclear fungal genome. We see the opposite of this which implies the Low coverage 4X contigs are likely from neighboring colonies or cannabis host DNA. If there were diffuse dead DNA, we would not expect them to form high coverage contigs but to represent a low coverage meta genome that would not assemble into pure high coverage long contigs. Usually the dead DNA is highly fragmented and diverse delivering fragmented, low quality assemblies. We see the opposite here.
The Table below has links to the Antibiotic resistance gene reports from OneCodex. This is a limited search of antibiotic resistance genes in the OneCodex database. These genomes are draft genomes. Coverage gaps and sequencing errors can limit detection of certain genes and the report is only as comprehensive as the database its is scanned against. Nevertheless, many antibiotic resistance genes are detected including resistance to the antibiotic in RYM plates.
| SAMPLE | AM-Resistance Report | # Antibiotic Resistance genes | AB Gene |
| 12283-1-5_001.fastq.gz | https://app.onecodex.com/panel/b9195357a2d24b42 | 1 | |
| 12335-1-3_001.fastq.gz | https://app.onecodex.com/panel/ca668fc742ca49f1 | 12 | Tetracyclines |
| 12335-1-4-LG_001.fastq.gz | https://app.onecodex.com/panel/98047a1c1b6c4304 | 9 | Tetracyclines |
| 12339-1-3_001.fastq.gz | https://app.onecodex.com/panel/6173d68f5d84490e | 9 | Tetracyclines |
| 12339-1-4_001.fastq.gz | https://app.onecodex.com/panel/d4cf17aae8bd4949 | 10 | Tetracyclines |
| 12352-1-1_001.fastq.gz | https://app.onecodex.com/panel/81710fa0ab7546b8 | 3 | Tetracyclines |
| 12352-1-5_001.fastq.gz | https://app.onecodex.com/panel/9d2f86063dcb4455 | 1 | |
| 12352-1-6_001.fastq.gz | https://app.onecodex.com/panel/b244a87bbf3e47a5 | 1 | |
| 12883-4_miseq.fastq.gz | https://app.onecodex.com/panel/52f29084a9bf41c5 | 0 |
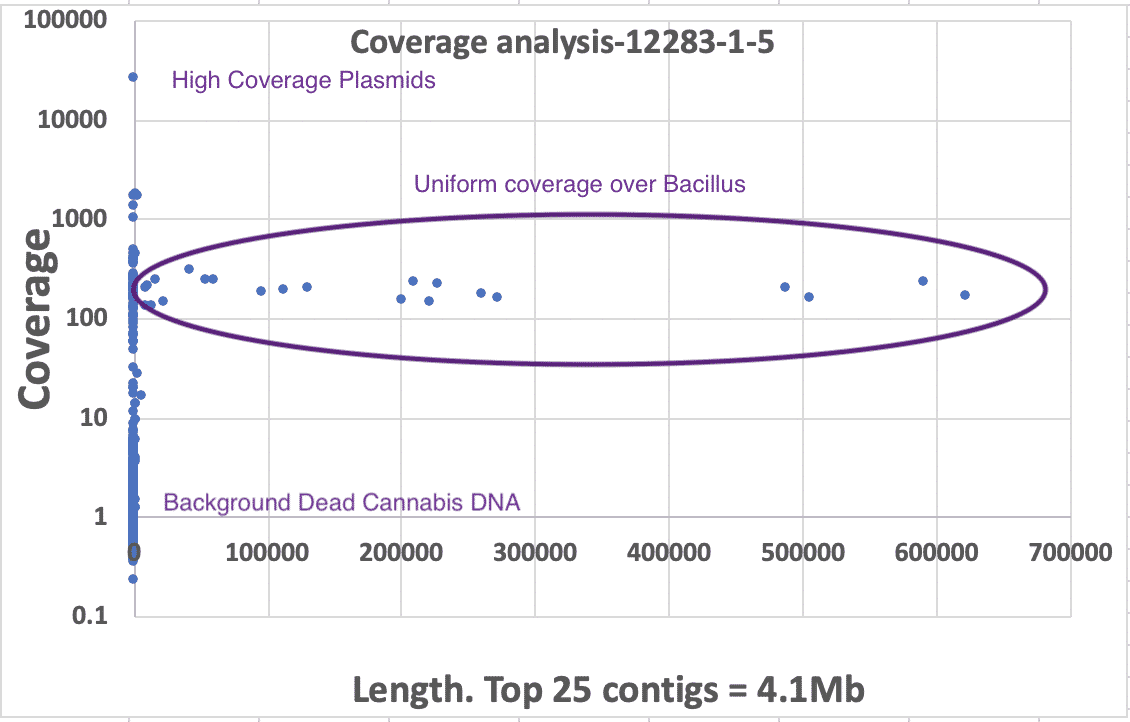
Presented in the chart above is the length of contigs on the X axis versus the sequence coverage of the contigs on the Y axis (log). One can see clear and uniform coverage for the Bacillus genome around 150X. High copy plasmids are short in length but appear high in coverage. Background dead DNA has low coverage and short contig lengths since dead DNA is highly fragmented. Cannabis DNA comprises the majority of the low coverage (~4x) short contigs.
We believe these demonstrate that the DNA isolated from colonies make up the majority of the assembly being used for analysis. Annotation of these genomes with OneCodex demonstrates multiple antibiotic resistance genes which agrees with the literature on these identified bacteria being resistant to the antibiotics used on RYM plates.
Other concerns were raised regarding the morphology of the colonies. Morphology is always secondary to DNA sequencing in taxonomy because many organisms exhibit pleiotropy. The classic case is caterpillars and butterflies. Different stages of the life cycle of an organism can present vastly different morphologies and thus morphology is never used as a primary proof of taxonomy.
An example of this with the Bacillus found in this study.
Living spores of B. amyloliquefaciens FZB42, now reclassified as a strain of B. velezensis, have been formulated into the commercially available bio-inoculant RhizoVital®, which is used to control a variety of soil-borne diseases https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6470737/
B. amyloiquefaciens and B. velezensis are now believed to be the same organism and they are endospore forming bacteria. These can form morphology that is easily confused with fungi. For this reason, the Tree of Life is organized by nucleic acid signatures not morphology.
To further emphasize this point, Bacillus amyloliquefaciens subspecies can be explored with many genome browsers to showcase the different genes and pathways these subspecies can contain. BioCyc contains many different subspecies genomes where one can easily compare CC178 with Y2. One can see there are 338 more genes in Y2 than in CC178. So not only can one not conflate Bacillus subtilis with Bacillus amyloliquefaciens but even conflating at the species level (Bacillus amyloliquefaciens Y2 with Bacillus amyloliquefaciens CC178) is known to be poor practice in the microbiology field.
Several Bacillus amyloliquefaciens products were acquired from grow stores, plated and showed no growth on RYM plates. It is unclear what subspecies was plated. Sequencing of these products is underway but the most effective solution to this problem is to maintain barcoded retains of the samples tested so people can go back and interrogate samples of interest. This does not appear to exist for the samples shipped to us from Michigan.
Independent of this, we can also look to Plate Count Performance data.
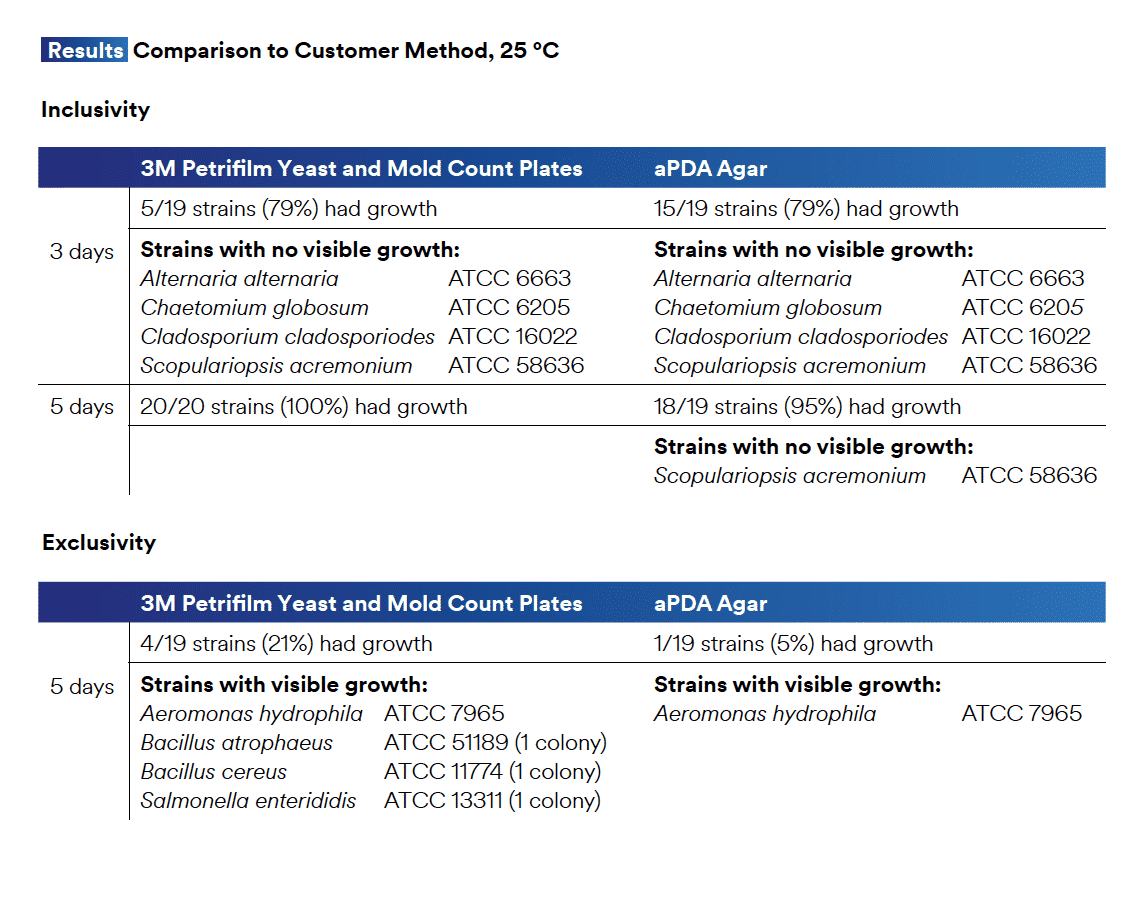
Likewise, one will note 21% of their bacterial exclusion organisms formed colonies on TYM and two Bacillus species are shown. This, once again, emphasizes the fallacy of conflating Bacillus genus and species and assuming the Yeast and Mold specificity in TYM plating.
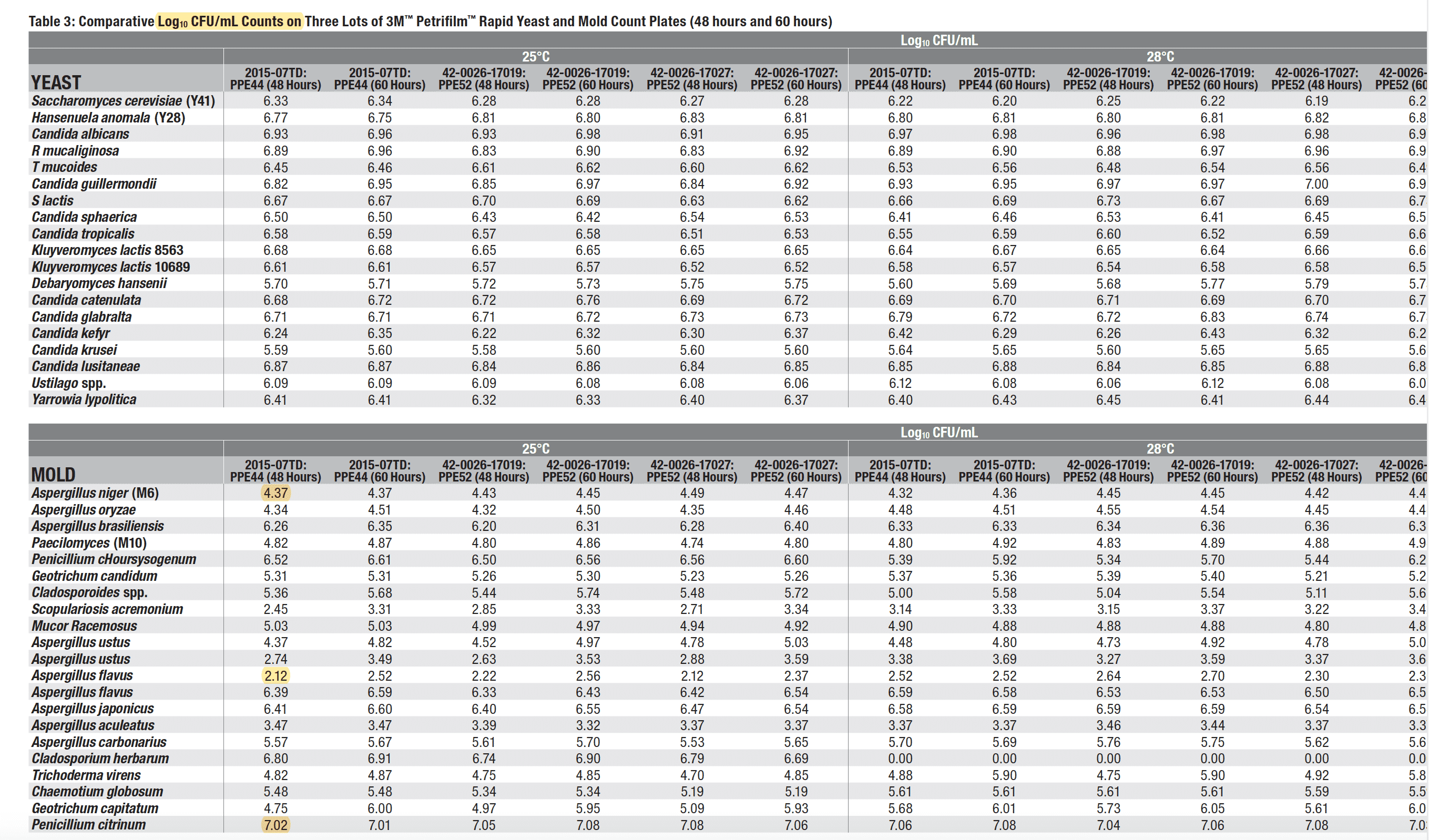
A further reminder of the complications in getting qPCR to match CFU/g can be seen in the validation data. Penicillium citrinum is known to be one of the most productively growing molds on plates with a Log10 CFU/g of over 7. Aspergillus flavus (which has killed cannabis patients) has a Log10 CFU/g of 2.12. This is close to a 100,000 fold difference in detection efficiency of two different molds tested. Most TYM regulations are set at 10,000 CFUs/g. If Aspergillus and Penicillium can vary this much, we don’t have a gold standard. We have an appeal to antiquity.
A link to these documents are here and here.
An alternative hypothesis requires mentioning. Since Bacillus has a faster doubling time than yeast and molds, it is possible to have a lawn of bacteria that is no longer an individual colony as a backdrop to fungal colonies emerging on them. In this case the bacteria may outnumber the fungi gDNA in DNA purification and dominate the sequencing data. Given its clear from the exclusion data that their plates can grow bacteria (Bacillus) and these plates were grown for several days, this can’t be ruled out. Very deep sequencing may reveal low levels of background fungal DNA.
We began itemizing the low coverage short contigs in the assembly and we can see Cannabis DNA (does not culture), Powdery Mildew DNA (does not culture) and some Penicillium DNA (cultures). Given the Penicillium contigs are at the same coverage and similar lengths to other background and unculturable organisms in the sequencing it is doubtful these represent the dominant the species sampled from the Colony.
The Michigan MRA sent out a letter reminding labs in Michigan that they must use validation organisms that are found on Cannabis. Unfortunately, the MRA assumed the organisms we used (Klebsiella pneumoniae) to validate our methods were not found on Cannabis. A Google search with the terms Klebsiella pneumoniae & Marijuana display Thompson et al as the top hit which clearly displays Klebsiella as a pathogen of concern on Cannabis.
The standard that presented this discrepancy was made from an organism (C.albicans) that has never been found on Cannabis and it appears from the MRA’s language in the below letter, it is no longer a suitable standard as there is no peer reviewed literature of this organism being found on Cannabis.
We applaud the MRA in bringing forward AOAC appendix J. There is currently no product on the market that meets these standards for TYM with a Cannabis matrix. We are working to validate the TYM assay according to AOAC appendix J. We have currently performed inclusion and exclusion criteria on over 140 organisms. RYM mold assay was validated with 40 organisms, none of which were selected to be from organisms frequently found on Cannabis. Its important to note the lack of specificity seen in this study where many bacteria grew on the plates.
It should also be noted that Appendix J section 6 is asking for 5 days of culture with no selection. The RYM tests run for 2 days with 2 antibiotics and do not meet this criteria. They will also have to be revalidated with a cannabis background matrix according to AOAC Appendix J.










