Culture-based methods, such as plating, are considered by some to be the “gold standard” of microbial safety testing. After all, the method is used extensively in the food industry and is validated on a myriad of different matrices.
However, cannabis is NOT one of those matrices! To date, there are no published manuscripts to support using culture-based plating methods on cannabis. In fact, our team published two manuscripts that point to massive clinical liabilities present with culture-based plating technologies. Most concerning is the fact that culture-based plating methods fail to detect real clinical risk. But even more practically, culture-based methods suffer from high false-positive rates.
Assuming methods that have been validated in the food industry apply to the cannabis matrix is a major error that can put consumers at risk. Anyone versed in cannabis testing can assure you, if you change the matrix, you must revalidate your technology. One cannot assume a method that works on tomatoes will work on cannabis.
Many of the top cannabis testing labs in the country use our DNA-based method for microbial detection, making it the industry’s preferred microbial testing technology. But some cannabis testing labs still use culture-based methods. Here are 5 reasons we believe they should stop.
Culture-based methods grow off-target species
As mentioned earlier, our a peer-reviewed article showed that over 60% of the DNA found on culture-based yeast and mold tests came from bacteria.
Most recently, we sent a saturated culture of an antibiotic-resistant Pseudomonas aeruginosa from ATCC to the two testing labs in Massachusetts using culture-based plating. We chose Pseudomonas aeruginosa because it is a Bile-Tolerant Gram-Negative (BTGN) bacteria, and the USP has listed it as required for any valid BTGN test. Massachusetts requires BTGN testing for all cannabis samples.
Both of the culture-based plating technologies used in those labs failed to detect the Pseudomonas aeruginosa in their BTGN tests. However, their Total Yeast and Mold test detected the Pseudomonas aeruginosa when it should not have. After all, Pseudomonas aeruginosa is a bacteria.
If these platforms cannot detect the proper organisms from a saturated culture without any cannabis present, you cannot rely on the results they give you with matrix or cannabis present.
Culture methods can’t detect harmful endophytes
Toxic Aspergillus species are responsible for at least one death of a cannabis patient. However, Aspergillus is an endophyte that lives inside the plant without causing apparent disease to the plant. Since most culture-based techniques rely on viable cells growing in a medium for detection, they do not lyse plant cells open to see the endogenous pathogens like DNA-based methods do. It is imperative that the flower samples be lysed so that the endophytes are accurately captured.
Even when Aspergillus does grow in culture, there are non-pathogenic Aspergillus species that look remarkably similar to their pathogenic cousins, making it difficult to speciate using visual identification alone.
In Alaska, there was a public case of misidentification where a lab mistook A. brasiliensis (non-pathogenic) for A. niger (pathogenic) and inappropriately failed the sample. A competing lab that uses our DNA-based method properly identified the species.
Only a fraction of microbes grow in culture, and even fewer grow well
Culture plating is a passive testing method. Labs place cells in a growth medium and hope it’s favorable enough for them to grow. The problem with that is only a fraction of microbes culture. This is known in the industry as the Great Plate Count Anomaly.
Furthermore, when multiple microbes are present in the same medium, they will compete with one another. Some species thrive in a given medium, while others grow very poorly. As a result, we have observed that the microbial population after culturing can be radically different than what is actually on the plant. See the image below.
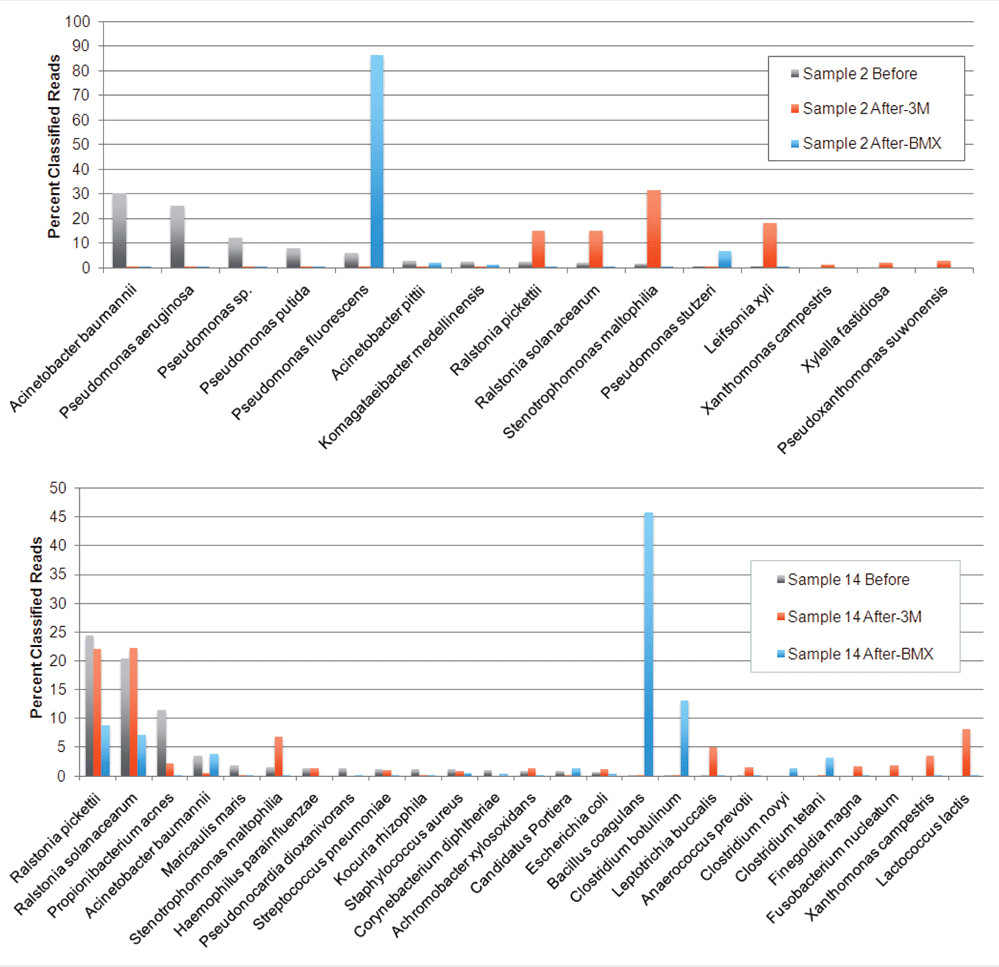
Culture methods fail in the presence of candies
Marijuana infused products (MIPs) are a very diverse class of matrices that behave very differently than cannabis flowers. Gummy bears, chocolates, oils and tinctures all present different challenges to culture-based techniques. The sugars and carbohydrates can radically alter the carbon sources available for growth. To assess the impact of MIPs on colony-forming units per gram of sample (CFU/g) enumeration, our team spiked a known amount of live E. coli into three different environments: tryptic soy broth (TSB), hemp oil and hard candy. The team then homogenized the samples, pipetted amounts from each onto Petrifilm E. coli / Coliform Count (EC) Plates, and incubated for 96 hours. The team also placed TSB without any E. coli onto a petrifilm to serve as a control. The images below show the results in 24-hour intervals.
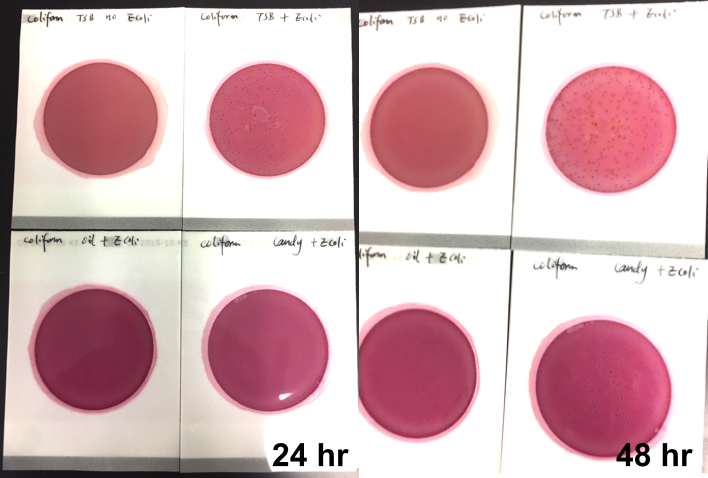
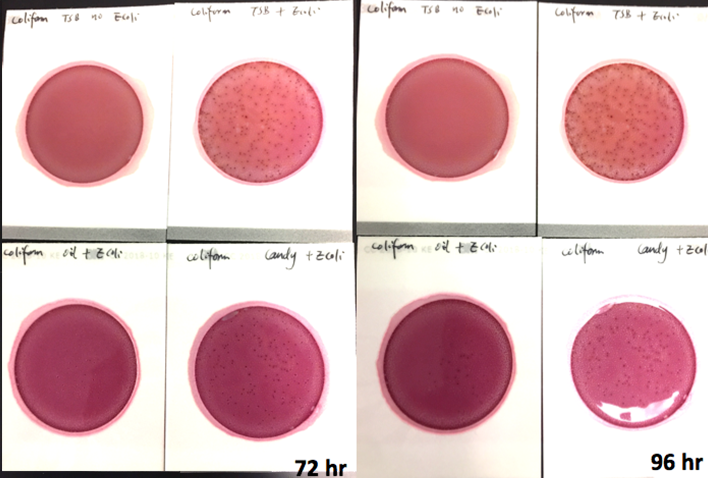
This implies the MIPs are either interfering with the reporter assay on the films or they are antiseptic in nature. Many MIPs use citric acid as a flavoring ingredient, which may be responsible for the interference.
qPCR is better
The Medicinal Genomics team completed an extensive manufacturer’s validation on the SenSATIVAx DNA extraction kit and PathoSEEK qPCR detection assays that demonstrates robustness, accuracy, and precision of the method on a variety of cannabis matrices. As a result, our qPCR solution addresses many of the problems with culture-based methods:
- qPCR assays actively look for specific DNA sequences shared by a family of microbes, making it impossible for off-target species to inflate results.
- qPCR assays are not subject to the Great Plate Count Anomaly because they do not rely on growth for detection. If the DNA is present, qPCR will detect it.
- This active approach means we can design species-specific assays that target the DNA signature for pathogenic species, such as Aspergillus, Salmonella, and STEC E.coli. No visual detection required.
- SenSATIVAx and PathoSEEK were specifically developed for microbial testing on cannabis flower, extracts, and concentrates and edibles.
It’s time we make qPCR the new “gold standard” in microbial safety testing.








