The list of problems with culture-based microbial testing methods continues to grow. The latest discovery from the Medicinal Genomics team found that four Enterobacteria species (Aeromonas hydrophila, Pantoea agglomorans, Yersinia enterocolitica, and Rahnella aquatilis) do not grow on common culture media when incubated at the manufacturer’s recommended temperature (36°C).
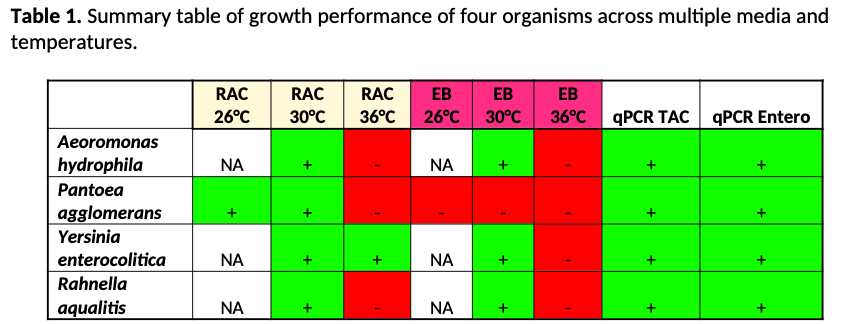
Aeromonas hydrophila, and Rahnella aquatilis failed to grow at 36°C on two different plating media (3M™ RAC and EB Petrifilms™) but successfully grew on these media at 30°C. Yersinia enterocolitica also failed to grow at 36°C on EB plates but grew successfully at 30°C on EB plates and grew at both 30°C and 36°C with RAC plates. Pantoea agglomerans only grew on RAC plates at 26°C and 30°C and did not grow on EB plates at any temperature. Conversely, qPCR detected all 4 organisms.
Pathogenicity and Prevalence
Even though these four organisms contain some similar DNA, they don’t carry the same pathogenicity, nor do they appear with the same prevalence on cannabis. For example, Aeromonas hydrophila is responsible for 13% of US cases of gastroenteritis and has been observed in cannabis at low levels. Pantoea agglomerans is less pathogenic, but much more commonly found in cannabis. Still, Pantoea agglomerans is known to cause skin rashes, which may be relevant for trimmers in constant contact with cannabis plant matter and sharp trimming tools.
Implications for validating alternative methods
Pathogenicity aside, this issue has implications for evaluating alternative microbial testing methods. qPCR is often critiqued as an inadequate replacement to culture-based enumeration methods because of its lack of concordance to plating. This becomes a circular argument when the plating methods have known and obvious blind spots. In this study, qPCR detected all of the pathogens and plating failed to culture all of them on 3M™ EB Petrifilm™ plates at 36°C and failed to culture 75% of them on RAC plates at 36°C. This is a scenario where qPCR might be accused of detecting non-viable organism when the organisms are, in fact, viable but just not viable on the chosen media or temperature.
Using a DNA-based testing technology is a far more universal and directed approach because it measures something guaranteed to be present in all organisms and that isn’t affected by variations in temperature, carbon sources or specific growth conditions. Conveniently, this universal analyte (DNA)is also the signature that classifies target organisms taxonomically. So for instance, bacteria can never be conflated with a yeast or mold at the DNA level. The use of this more specific, universal analyte is the path to future lab-to-lab, and state-to-state, concordance. Further evidence of that is its certification by AOAC for Aspergillus and SAL/STEC for cannabis flower and edibles.
None of this, of course, takes into account the risks to laboratory personnel, who have to work with these human pathogens, which is not inconsequential. Still, the greatest risk to cannabis patients and for that matter, the industry, is the idea that reliance on a hundred and thirty-year-old plating “technology” from the food industry will work just fine on cannabis. It won’t and it doesn’t. And while cultivators would rather have labs use methods that pass more cannabis than they fail, the consequences of pocketing higher profits, while incurring greater risks serves no one—not the reputation of the industry at large, not cultivators, not labs, and certainly not patients—in the long run.
Implications for Growers
For the growers concerned about picking up organisms their current plating methods don’t but should detect, we would encourage you to look at our previous peer reviewed publication “Whole genome sequencing of colonies derived from cannabis flowers and the impact of media selection on benchmarking total yeast and mold detection tools”. In this study, three different plating media were compared and showed log scale differences in the number of colonies detected. One Total Yeast and Mold media (PDA [potato dextrose agar]) contained 90% bacteria. If you are attracted to plates that under-detect specific organisms you have to make a devil’s bargain that erodes your specificity. The path to picking up fewer CFUs of a specific microbe, may in fact expose you to picking up more off-target colonies, leading to false failures. It is very difficult to build a predictable business just from “reading tea leaves” in the final QC step.
For all the details on this story, including our methodology, results and sources, visit https://osf.io/j3msk/, on the Center for Open Science’s website and the transparent review/open access journal F1000Research https://f1000research.com/articles/10-624