Earlier this week, we made some exciting updates to Kannapedia, the website that houses genetic data from the cannabis cultivars sequenced with our StrainSEEK® Strain Identification and Registration Service.
We added new data visualizations and genetic distance tables that make it easy to compare cannabis cultivars in the Kannapedia database. Cultivars that we sequenced with our StrainSEEK® Version 2 panel now also include a table that compares them to cultivars published to Phylos Bioscience’s Open Cannabis Project.
We can do this because we designed the assay to include loci from the other public cannabis genomic datasets (Phylos, Lynch, Sawler). StrainSEEK® is the only cannabis strain sequencing service that can do this.
Read more about the updates below.
New Phylotree
The targeted approach we are using for StrainSEEK® delivers 25,000-50,000 SNPs across the genome with a concentrated contribution from chemotype related genes. The higher SNPs density enables Marker Assisted Selection.
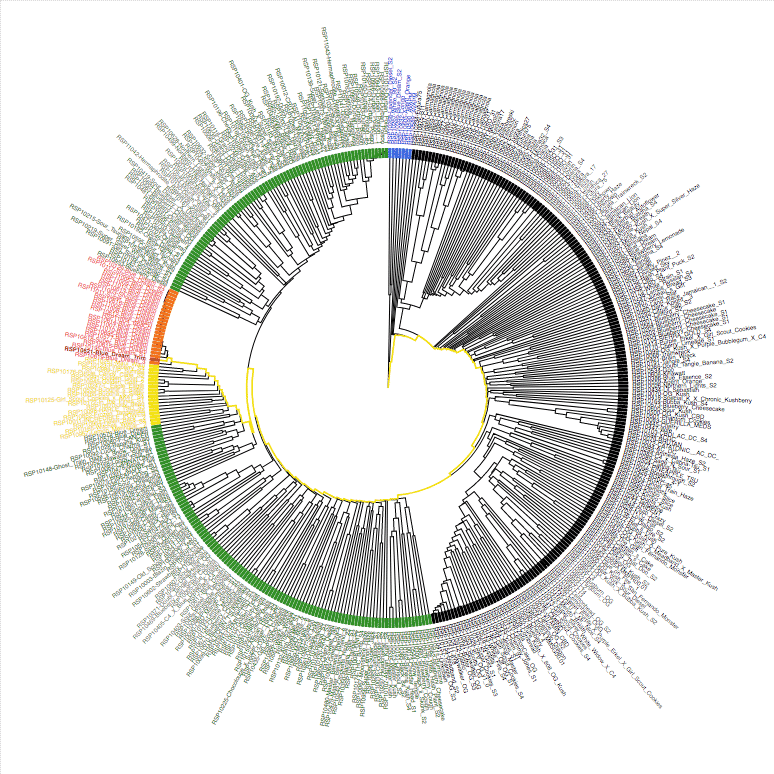
This phylotree is derived using the intersecting high-quality SNPs from StrainSEEK® V1 and StrainSEEK® V2. The SNPs with the higher information content in StrainSEEK® V1 were carried over into StrainSEEK® V2 for backwards compatibility during the upgrade.
Strain Rarity Visualization
Each strain page now includes a Strain Rarity Visualization, which shows how distant the cultivar is from the other cultivars in the Kannapedia database.
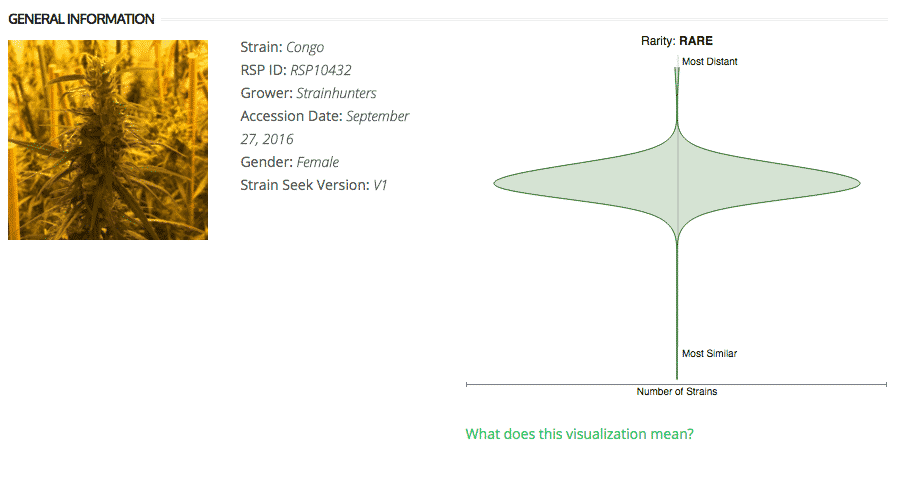
The y-axis represents genetic distance, getting farther as you go up. The width of the visualization at any position along the y-axis shows how many strains there are in the database at that genetic distance. So, a common strain will have a more bottom-heavy shape, while uncommon and rare cultivars will have a visualization that is generally shifted towards the top.
Heterozygosity Visualization
Each Strain Page also shows how the cultivars heterozygosity compares to others in the database.
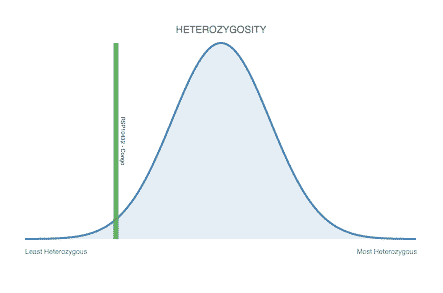
The bell curve in the heterozygosity visualization shows the distribution of heterozygosity levels for cannabis cultivars in the Kannapedia database. The green line shows where this particular strain fits within the distribution. Heterozygosity is associated with heterosis (aka hybrid vigor) but also leads to the production of more variable offspring. When plants have two genetically different parents, heterozygosity levels will be higher than if it has been inbred or backcrossed repeatedly.
Most Genetically Distant Strains Dot Plot
We added a new table and dot plot which shows the 10 cultivars that are most distant.
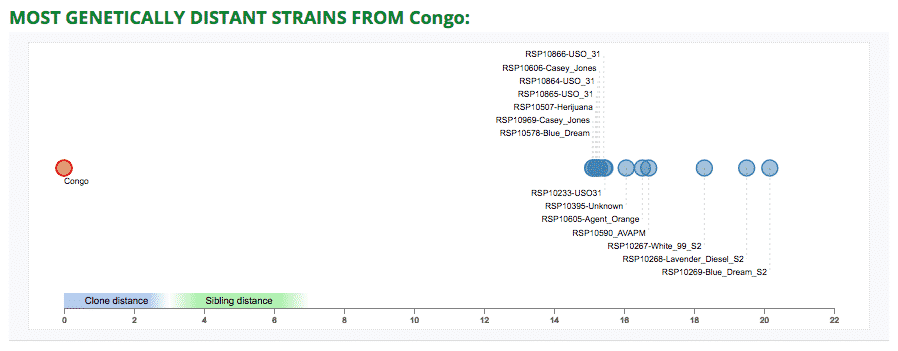
This can be useful information for breeders who are interested in creating new and interesting cultivars. Breeding with genetically distant cultivars will produce diverse phenotypes, which will likely have high heterozygosity or hybrid vigor.
Nearest Genetic Relatives in Phylos Dataset
Finally, we added a table that compares genetic data from cultivars sequenced with the StrainSEEK® Version 2 panel to cultivars that were sequenced by Phylos and published to the Open Cannabis Project.
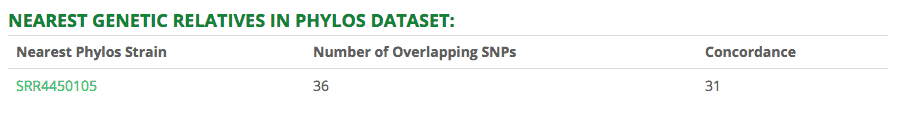
We can do this because we designed the assay to include loci from the other public cannabis genomic datasets (Phylos, Lynch, Sawler). StrainSEEK® is the only cannabis strain sequencing service that can do this.
As result, samples sequenced with this method can be cross compared to all data that is public as of 2017. StrainSEEK® version 2 delivers more than 10 times the genetic information than other tests on the market. and as a result is the most comprehensive sequencing tool. More data means you can better protect your strain from patents, better determine if your strain is unique, better identify genetic markers for breeding, and much more.