We added four new features to the genomic services reports on Kannapedia.net:
- Cannabinoid and flowering genes added to the variant table (40 genes curated by the Medicinal Genomics team)
- Variant frequency information added to variant tables
- Integrative Genomics Viewer (IGV) integration
- New Downloadable Files
New Cannabinoid and Flowering Genes in the Variant Table
Previously, Kannapedia included one variant table showing any point mutations determined to be high impact variants for THCA synthase (THCAS), CBDA synthase (CBDAS) and CBCA synthase (CBCAS). We believe these three cannabinoid precursors are likely to be of the most interest to users, which is why they were pulled out and highlighted in the report.
Now we are highlighting other genes of interest that are likely to play a role in Cannabinoid synthesis and flowering. A complete summary of the genes can be found here. Key features of the new table are described below.
High impact variants are variants that are more likely to change the amino acid sequence of the protein and are thus more likely to alter the function of the protein for a given sample.
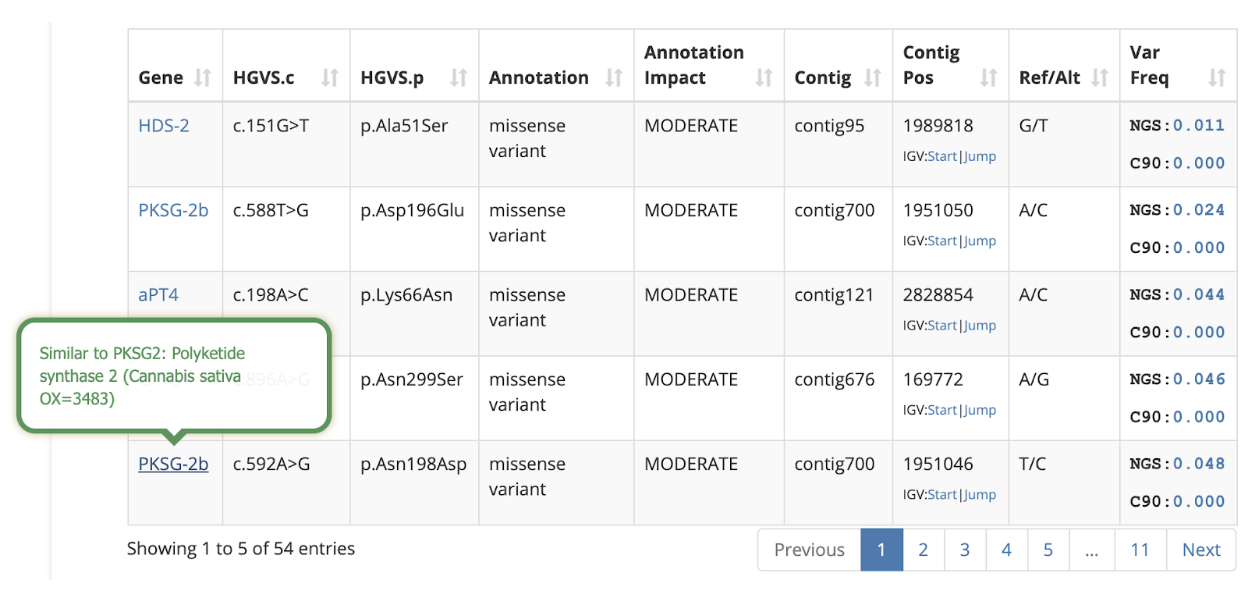
Variant Frequency Information
Some variants are common and are shared among the population. These are of less interest since any common variant is less likely to affect anything about the protein for which the DNA encodes. On the other hand, rare variants could cause unique mutations that could alter a cultivar’s phenotype.
The variant table now includes a Variant Frequency column that shows the frequency of each compared to other cultivars in the database.
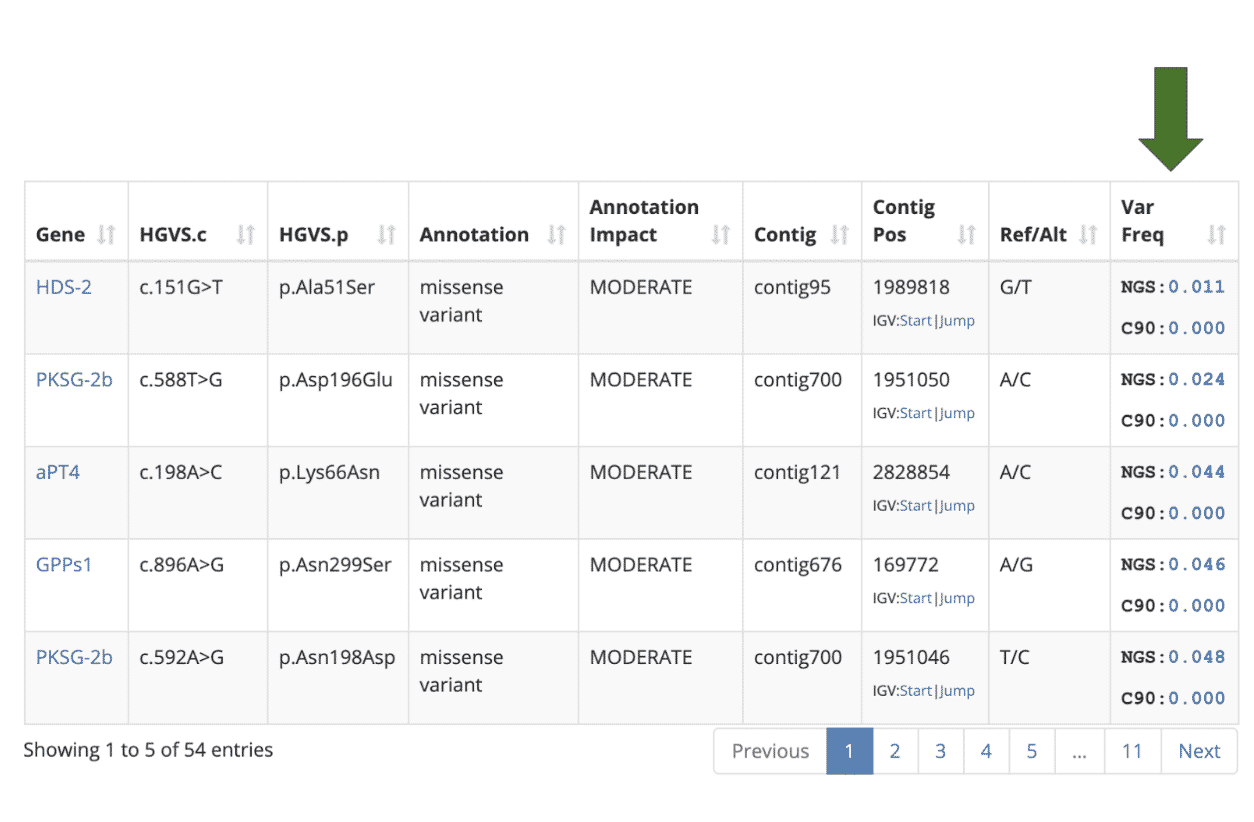
The “NGS” value shows how common the variant is among samples that were analyzed with Next Generation Sequencing technology (i.e. StrainSEEK® or Whole Genome Sequencing). The “C90” value shows how common the variant is among samples that were analyzed with the CannSNP90 Chip. For example, if there are 1000 samples in our database and the NGS Var Freq column indicates 0.100 then this variant is present in 100 (10%) of the NGS samples in the Kannapedia database.
By default, the variant table is sorted to show the lowest frequency variants from the NGS data sets, assuming they are potentially the most interesting. Hovering over any of the variant frequency values will show a list of samples that share the variant. The example below shows five other NGS-based samples in Kannapedia that share the first variant in the table (p.Ala51Ser in the HDS-2 gene). The five RSP identifiers are shown along with clickable links that take you right to those reports.
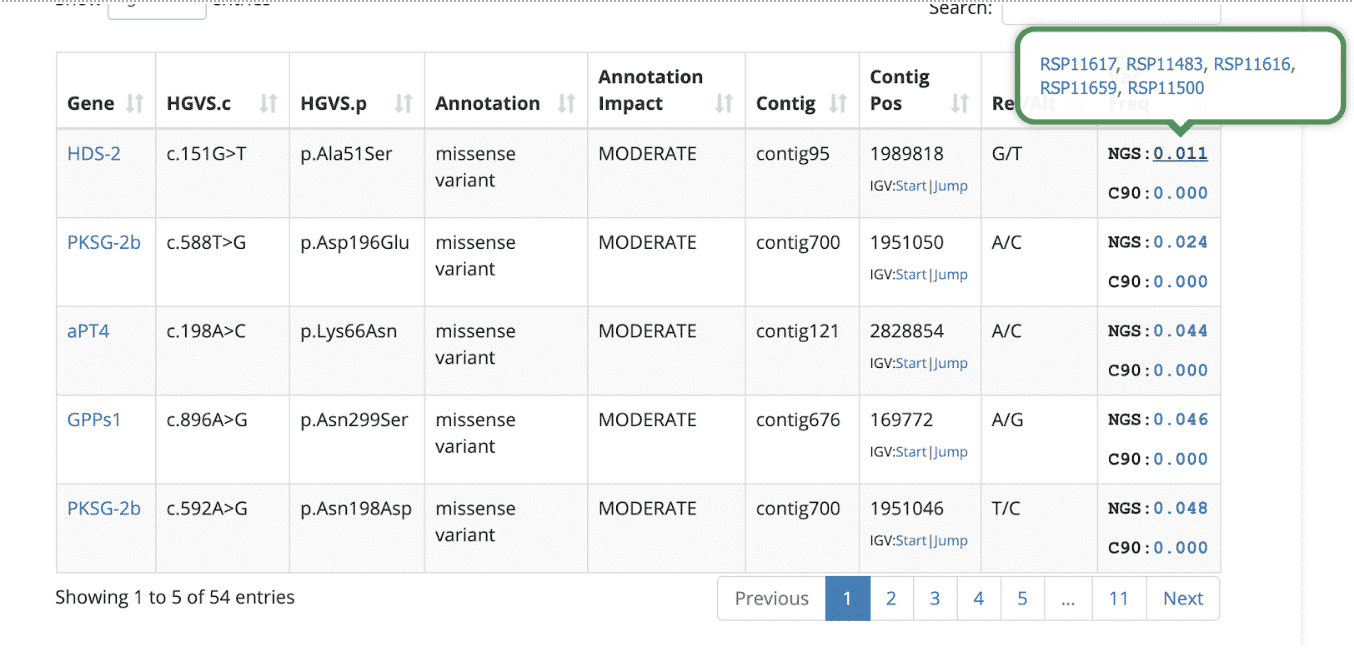
Hover over the gene name to read a short description:
IGV Integration
The Integrative Genomics Viewer (IGV) from The Broad Institute allows users to view sequencing reads and variants in a graphical way that can be insightful.
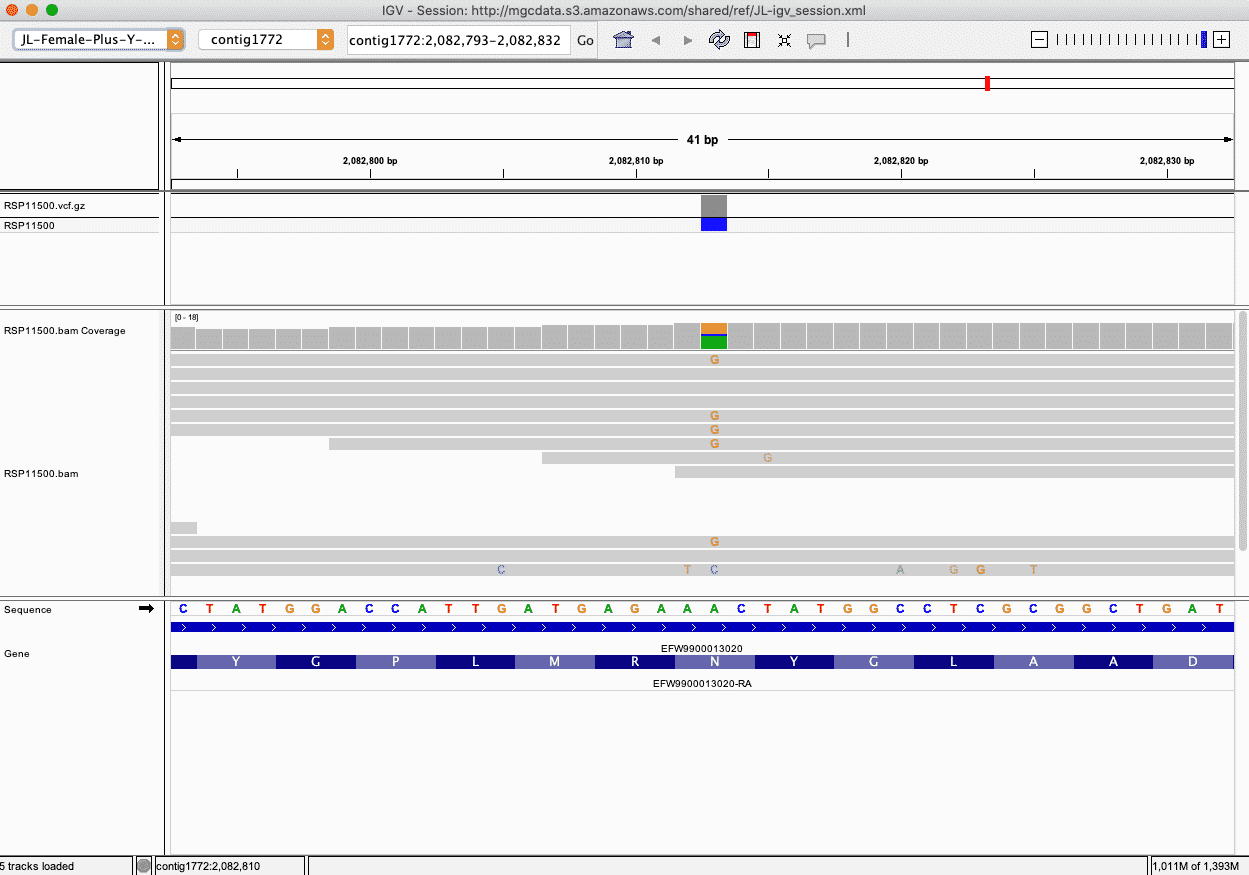
Variant information for each genomic services sample is stored in a VCF file. Additionally, NGS-based samples have mapped sequencing reads stored in what is called a BAM file.
The IGV shows the variant information and the sequencing reads (if applicable) mapped to the Jamaican Lion Cannabis reference from Medicinal Genomics, which is stored as a FASTA file along with gene annotation in a GFF file. IGV does the work of indexing all these files so that the user can quickly jump from one position to another. Using IGV, you can load any particular region of interest (not just these variants) in the Cannabis genome and see all of these genomic features presented in a visual way.
Users have to install IGV on their computers in order to use its features. Read our guide to getting started with IGV
New Downloadable Files
SNPEff-annotated VCF files are now available for NGS and CannSNP90 samples. BAM files are also available for any NGS-based Kannapedia reports.
Links to downloads these files can be found in the GENETIC INFORMATION section of the Kannapedia report.

As these are binary indexed files, an index file is also provided for each in the accompanying index link. The index files are the key to being able to use NGS-analysis tools like samtools and bcftools to quickly access any position in the Cannabis genome.
To best make use of these files and these tools, users must download the reference genome that matches the BAM and VCF files. Download the Jamaican Lion reference genome in FASTA format and its index file using the links below