Hop Latent Viroid (HpLVd) is as enigmatic as it is ubiquitous. Although one study estimates as much as 90% of California cannabis is infected with HpLVd, costing nearly $4 billion in lost yields, the viroid’s pathological mechanism in Cannabis sativa is still unknown.
A few studies in closely-related hops (humulus lupulus) suggest that HpLVd may exert its pathology through RNA interference, which occurs when short viroid sequences interfere with the expression of genes with which they have sequence homology.
To better understand HpLVd pathology and potentially identify which genes HpLVd could interfere with, our team:
- Downloaded all 162 HpLVd genomes from NCBI
- Aligned the HpLVd genomes against the C.sativa (Jamaican Lion) transcriptome
- Annotated the short sequence homologies identified to the various genes
- Aligned all viruses in NCBI, including those not known to infect cannabis to the cannabis transcriptome as a reference point for off-target homology.
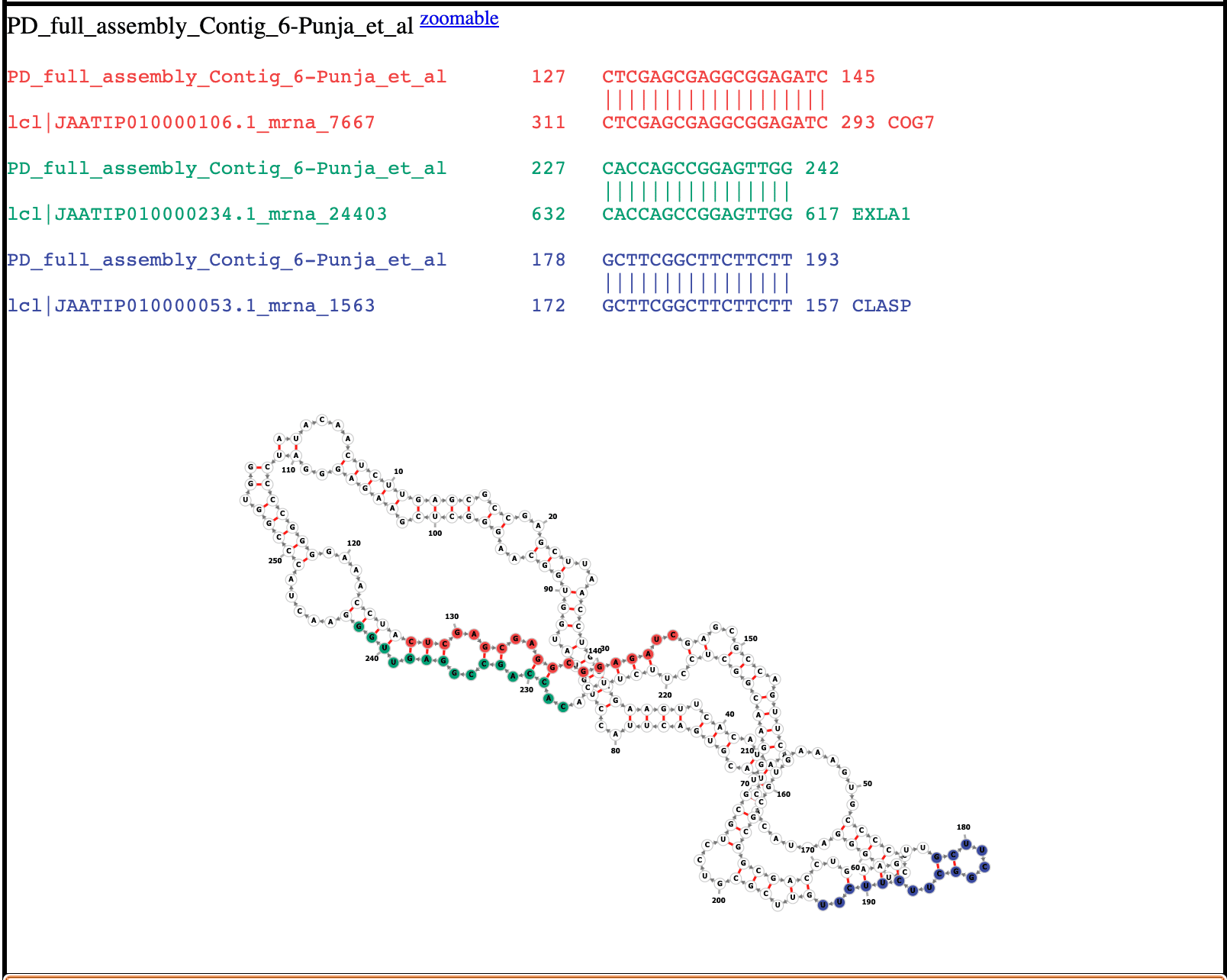
This process identified a few candidate genes for RNA interference. The most notable candidate is the COG7 gene, which is involved in the formation of the organized shoot apical meristem. So far, we have found that 69% of the published HpLVd genomes have homology to the COG7 gene, which may explain how HpLVd affects plant growth. There are 24 other genes in the cannabis genome that have short homologies to HpLVd genomes described in our latest preprint.
What does this mean for growers?
We are actively looking to work with growers and labs to further understand the COG7 pathway as we believe it is the only path of the qPCR treadmill. We need to start breeding for plants that are more tolerant of this viroid, and it is possible that cultivars with certain variants within the COG7 gene could be resistant to RNA interference caused by HpLVd. It is important to distinguish tolerance to HpLVd infection and resistance. In the case of evading the RNA interference of the viroid, the plant will not halt the replication of the viroid and thus still remain PCR positive. We also know that some HpLVd genomes have homology to a cannabis Callose synthase gene (CALS8). This gene is involved in plant cell porosity, viroid trafficking, and plasmodesmota formation and has been documented in PSTVd pathology in tomatoes. Only a select few HpLVd viroid genomes have mutations that target CALS8 in cannabis. This underscores the need to know your viroid genome and know the genomes of your mother plants.
We know that HpLVd has other plant reservoirs and reservoirs that may also include fungi. Complete elimination of HpLVd is unlikely. It hasn’t occurred in Hop, and that field has had many more years of experience dealing with the viroid. The first Hop Latent Viroid was published in 1988, and it is still a problem today for some cultivars more than others.
What are we doing to solve the problem?
We designed a qPCR assay to monitor COG7 expression in infected and non-infected plants. Preliminarily, we see a change (delta CT) but the plants in this study were not clones of each other and this work needs expansion into more cultivars.
We noticed five cannabis cultivars in Kannapedia.net have mutations in this region of COG7. These mutations may limit the capacity of HpLVd to knock down the expression of this gene. Further work is ongoing to catalog all of the cannabis variants in the 1,777 samples in the Kannapedia database and their impact on all HpLVd sequences known to date.
To facilitate this, we built Viropedia.net, which visualizes the secondary structure of all HpLVd viroids published to date and their respective homologies to the Jamaican Lion genome.
We’d like to thank Zamir Punja and Collin Palmer for submitting additional HpLVd genomes to this public resource.
Since the HpLVd genome is very polymorphic, there is a surprising amount of diversity in the cannabis genes it shares homology with. Our preprint describes 24 different cannabis genes that share sequence homology with HpLVd genomes.
We are actively sequencing HpLVd genomes and Cannabis genomes submitted by growers to sort this out. Contact us if you want to build a long-term plan that can greatly augment the qPCR screening currently being deployed.







