Until 2017, DNA sequencing cannabis strains has been a task only accessible to the well banked. The cost of DNA sequencers were $100,000 – $1,000,000 and the ancillary equipment required to prepare samples for such a task would add another $50,000 to the infrastructure costs. While many labs would accept DNA sequencing for a service, they would NOT accept cannabis samples in their doors.
This left scientists with clandestine DNA isolation options that could be hacked in hotel rooms however most companies that sell these traditional biotech kits and tools would not ship their kits to a residential address. This is part of the story of the first cannabis genome sequenced in 2011 (Chemdawg) in the Dylan Hotel in Amsterdam.
2017 is here and Oxford Nanopore Technologies is prepared to disrupt the field with USB stick sequencers that are under $1000. These sequencers produce very long reads (over 10Kb) but are only 90% accurate. This will create a point of grow genomics market and this community page is a resource to this end.
This page is dedicated to Sequence data derived from an ONT MinION data. Please join our email group and facebook page for more collaboration on this project. We are experimenting with assembling this data with Canu and Spades but encourage others to join the Cannabis Nanopore Swarm and help make a better public reference. Feel free to download the data below. Any derivative works we ask to remain open source under creative commons share alike.
We will be highlighting more of this project at CannMed2017. Please Join Us!
50Kb library Test Set on R9.4 1D Flow cells with MinKNOW v1.4
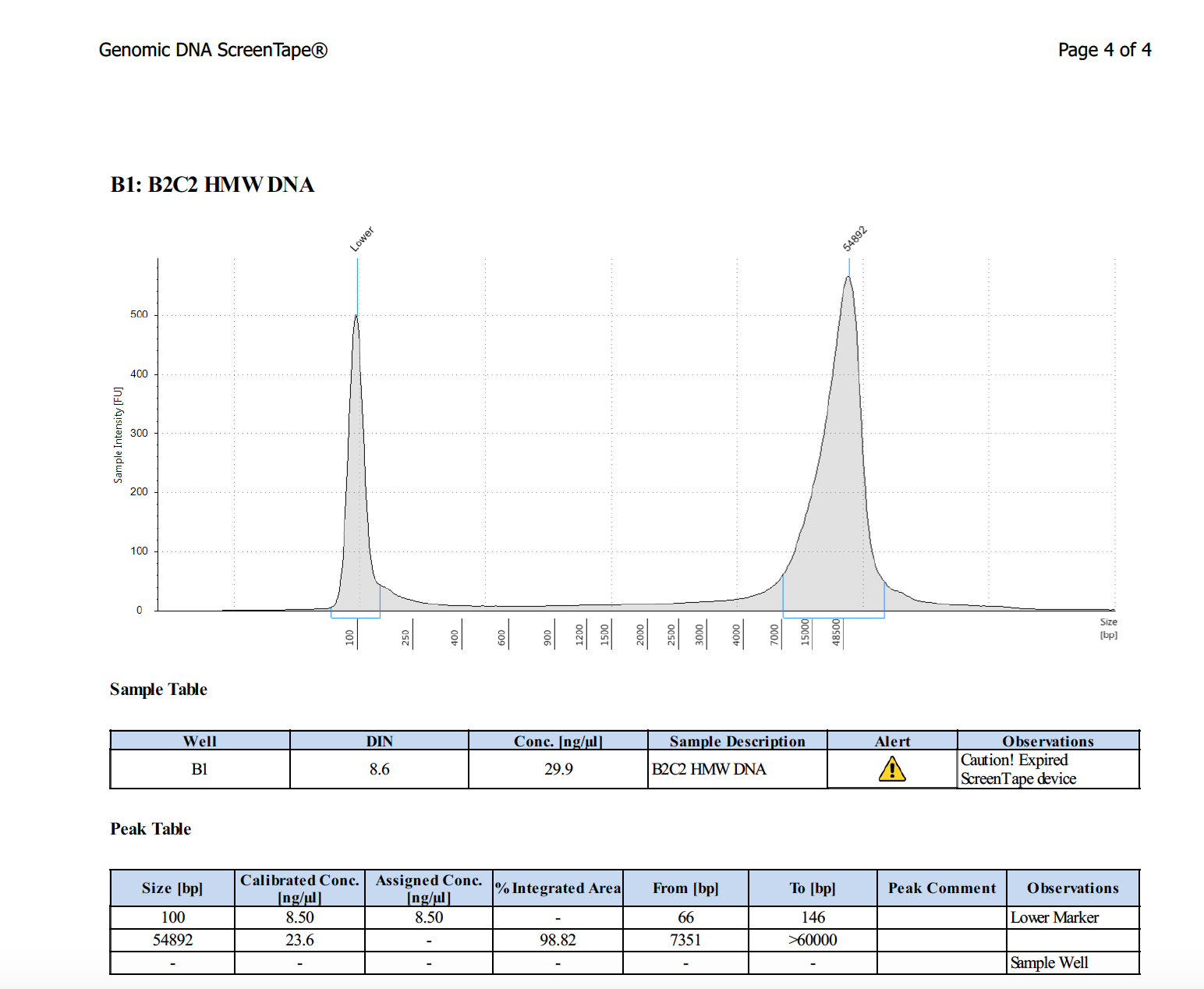
DNA was prepped using a CTAB method optimized for High Molecular Weight DNA (HMW).
End Repair and Adaptor Ligation are required before Nanopore loading (~2hrs).
Oxford Nanopore Sequencer
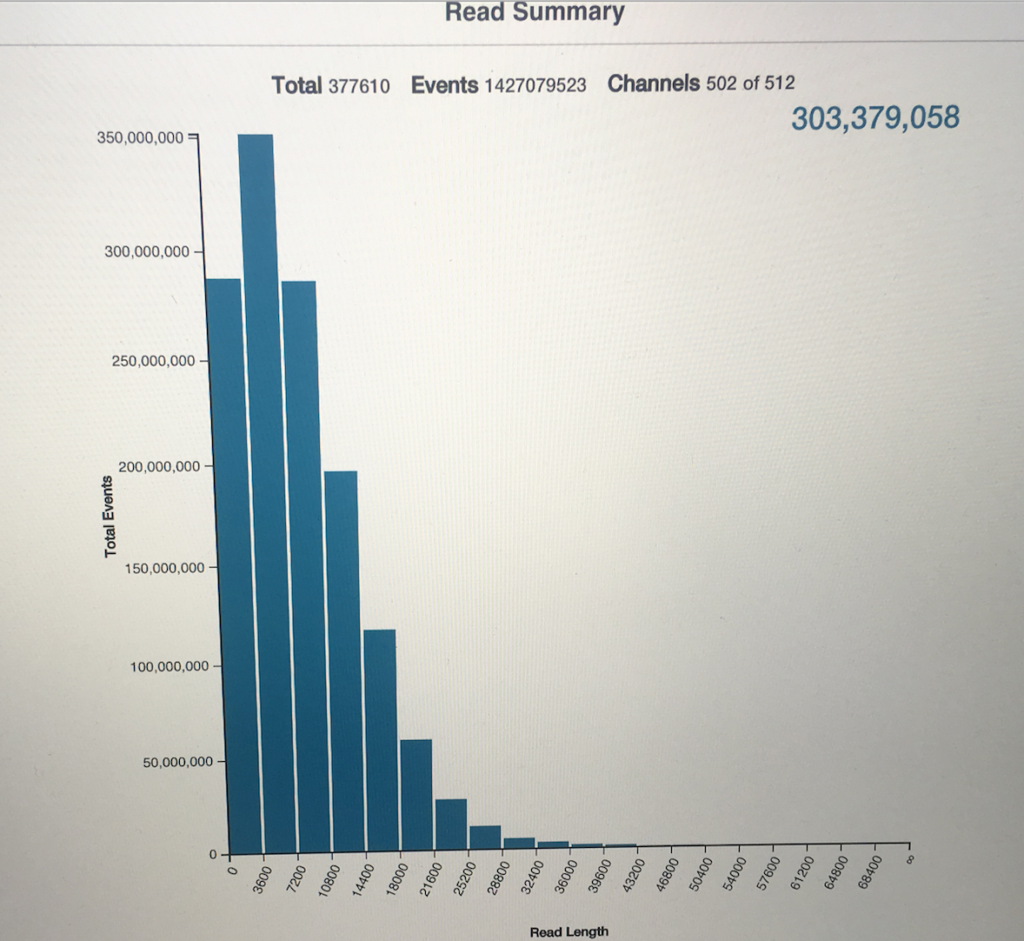
Read Histogram from the MinION