There are two important reasons to expand this investment.
1)Very Broad Cannabis Patents are now issuing. Any plant that makes CBDA and THCA are now covered by this patent. These are called Type II Cannabis plants.
Many people claim to have Type II plants still alive today that predate the 2013 filing of the patent. In fact Type II plants are believed to be the natural Hardy Weinberg equilibrium of the THCA/CBDA synthase genetics. Humans bred toward Type I plants during prohibition. Type I plants synthesize all THCA and little to no CBDA. Fortunately we published the genome of a Type I plant in 2011 to assist in prior art generation and public ownership of Type I plants. By placing the sequence of a Type II plant public, we can help enable the community to sequence their documented Pre-2013 Type II plants. They can then compare their sequence to this public reference to document the Bt:Bd allele (mentioned in some of the claims) and any other similarities. This will assist in building a Prior Use Exemption and more public evidence for the invalidity of the claims. Contributors to this project are published on the topic of gene patents and known for devising methods to weaken them.
2)Peer Review is broken and DASH can fix it. 50% of peer reviewed Scientific manuscripts describe experiments that can’t be reproduced. The journals with the higher impact factor have a higher retraction rate. Retraction rates are increasing dramatically. The Peer review incentives are based on costly and rusty Gutenberg era publishing economics. There is a near zero cost to publication on the internet today. Crypto-incentivized, blockchain transparent peer review will dis-intermediate this market and re-align the incentive models for higher accuracy, more transparency and faster turn around times. A simplified economic model is presented HERE.
We have placed a public crypto address for people outside of the DASH MNO network to contribute to the project.
We have chosen to do this exclusively with DASH while the project is DASH funded. We may open this to alternative coins after the DASH funding period. For a detailed scientific dissection of the project, please see the DASH proposal above.
XxXdB9HZ4gTCZNUGV5g6rj2eT8CgnL16mv
Thank you for the consideration and public charity.
If you would like your name to be on the attribution list of the genomes landing page please contact us at info@medicinalgenomics.com. It is possible to link your name to a donating crypto address via digital signature.
Shortly after Ernest Hancock covered this topic on his Radio Show, Elon Musk tweeted a similar idea. He is late to the game behind Trive.news who has something like this currently running to fight fake news. It is our belief that one can not fix fake news without fixing fake science. If 50% of the scientific papers can not be reproduced we need to extend these Trive.news concepts to scientific peer review. The scientific content requires data stores far larger than news articles and thus we are encouraged by the architecture behind DASH drive.
DASH has officially Funded the Project!
Strain Selection Criteria:
1)The plant should have a Type II DNA confirmation of the Bt:Bd allele using youPCR and methods described by McKernan et al.
2)The plant should have a history that predates 2013.
3)The plant should be currently alive and in circulation to enable further study of the plants genetics.
4)The plant should have low heterozygosity to assist in genome assembly.
We screened candidate cultivars with Medicinal Genomics youPCR THCA and CBDA assays to find Type II plants.
June 4th, 2018
DASH Hash of image below-
8cecc2636272ecc2630e565c389d34a8a82d75930c2d3e93342e6f7179373beb
Long Range PCR was also performed on THCAS and CBDAS. DNA was purified and run on an Agilent Tape Station to verify the youPCR colorimetric assays.
DNA isolations for Illumina Whole Genome Shotgun Sequencing are under way. Ordered 0.75% gels from Sage Biosciences for 40Kb selections of DNA for Pacific Biosciences sequencing.
June 5th, 2018
DNA purification for Pacific Biosciences using modifications of the Medicinal Genomics MIP DNA purification kit. DNA will require additional size separations to eliminate the long tail of small fragments of DNA.
DNA was also purified and fragmented for Whole Genome Sequencing using an Illumina MiSeq. DNA was tagmented with Nextera and size selected on a Blue Pippin.
June 7th, 2018
Post Size Selected Library ready for Loading on a Miseq for 2×250 bp reads.
MiSeq Cluster Density on the Hi side:) We will have to revisit the quality after the strand flipping. This tends to increase the diameter of the clusters and may make the reverse reads less legible.
DASH Hash of image below-
e7ef91922ea6cdc87fe9b557791a9e8d643d2c5a9e121fb7f2bb1f4d95409b13
Pseudo-colored Fluorescent images of DNA clusters on MiSeq Flow Cells.
July 8th, 2018
The strain we have selected for sequencing is Jamaican Lion^3 from Emerald Pharms and Ganjahnetics. Since this is from a seed, its is not a clone of the historic strain that has won cannabis cups. It is a descendant and thus genetically related but unique. We have “coined” the name “Jamaican Lion^3 DASH” for this offspring.
The History of this strain dates back to 2007. The Biotech LLC patent has a 2013 priority date. Jamaican Lion even came in 2nd place in a 2011 San Francisco High Times Cannabis Cup. Steep Hill labs was instrumental in identifying CBDA in samples like this which predate the patent. Steep Hill labs was the first cannabis testing facility in California and probably the world. This is an excellent example of how free markets even in the face of prohibition still value safety and quality.
The below chemotype report from Sonoma Labs is dated in 2016. It is from of a plant grown from a seed from the same mother. Its is not the identical plant but a sibling of what we are sequencing today and it matches what is known from other sources about this 2007 plant.
A second Medium Molecular Weight DNA prep was performed to obtain enough material to afford a size selection for 30Kb+.
400ul of 39ng/ul DNA was obtained or just over 15ug of DNA. A close examination of the electropherogram below will reveal lots of lower molecular weight DNA with a peak of the mass at 20Kb. The smaller DNA clones preferentially. As a result a size selection of the DNA is required to continue sequencing with the Pacific Biosciences platform.
June 9th-
Downloaded 7Gb of Illumina MiSeq 2x250bp read files. Trimmed Nextera adaptors with CLC Bio. Upload Trimmed Fast files to MEGA.nz.
These are ~3.6Gb & 3.9Gb gzip fasts files.
June 10th, 2018
Map reads to reference genome (Jim Knight’s LA Confidential Reference ) to assess insert size and quality. Confirm Type II status. May need to FLASH the reads to capture overlapping pairs.
Mapping Statistics:
Playing with FLASH to unite the Forward and Reverse reads. Since 250bp paired reads on inserts under 500bps will overlap, FLASH can combine them into one synthetic 400-500bp read. 86% of the reads can be joined by FLASH (using -M 240 -m25 on v1.2.11). Mapping these to CanSat3 to see if the broken paired rate can be improved.
June 11th-2018
Mapping FLASH’d reads to THCAS reference sequence with 98% Length Fraction at 98% similarity fraction with CLCBio V9.0.1 demonstrates a fully functional THCAS gene or Bt allele. Green reads are mapping on the Watson strand and red reads are mapping on the Crick strand. Since the reads were FLASH’d together, there are no paired reads.
Similar read mapping analysis on CBDAS reference sequence displays a functional gene. We suspect there are 2 functional Bd alleles. There may only be one functional Bt allele but Pacific Bioscience sequencing is a requirement to perfectly phase this region of the genome. Further chemical analysis should be completed before Pacific Bioscience sequencing begins.
June 12, 2018
Kannapedia.net places this Type II strain closest to Blueberry Cheesecake. While these are excellent Type II candidates for sequencing, the heterozygosity of the strain is excessive (3.4%) and not ideal for De Novo sequencing. Further screening of Type II plants with a faster and more scalable StrainSeek (1Gb panel vs 8Gb Whole Genome Shotgun) will be used to identify a lower heterozygosity Type II plant for Pacific Bioscience sequencing. The heterozygosity should not be taken literally. This is a metric that is measured on mapping to a compromised reference genome and is likely inflated due to mis-mapping of reads. It is also not a genome-wide measurement. It is a measurement of a BED file that covers 3.2Mb which includes 30 genes under the highest selective pressure. Many of these genes have been published to have aneuploidy or Copy Number variations and thus have inflated heterozygosity. It is possible Jamaican Lion parents could produce a wide variety of Type II offspring that are close to Blueberry cheesecake. Given we were screening Blueberry Cheesecake genetics simultaneously, we cannot rule out a sample swap and should continue to screen more seedlings.
June 13, 2018
Robotic Isolation of Jamaican Lion #2 and Jamaican Lion-Yellow DNA from fan leave stems. Barcoded robotic DNA purification greatly reduces the potential of sample swaps.
June 14,2018
More youPCR screening for Type II plants was performed. Both candidates are positive for a functional CBDAS gene. Concern over Botrytis infections polluting the shotgun assembly was raised. Plant is also negative for Botrytis according to MGC youPCR Botrytis assays.
Two StrainSEEK V2 libraries were made to sequence and check for heterozygosity. These will be sequenced on an Illumina MiSeq and uploaded to Kannapedia.net.
We initiated discussions with various Massachusetts growers to house and propagate the selected plant and to consider starting a tissue culture repository. This would make the first Cannabis genome reference that reflects a living plant where a researcher could acquire a cutting or a seed of the plant. This is very important for building a scientific reference standard. It is a critical resource for projects looking to utilize RNA interference as a pest control strategy. It is critical for any CRISPR/Cas9 projects looking to design targets for capture. It is also critical to ensure others can reproduce this work.
June 15,2018
DNA samples for JL#2 and JL-yellow were prepared for Agilent SureSelect Capture.
Caught up with DASH team members on the project.
June 18th, 2018
StrainSEEK Hybridization Capture ran from Sunday to Monday. Given these 2 samples pass QC, it will graduate to a MiSeq run on the 19th. Sequence data should be processed by end of week.
Medium Molecular weight DNA purifications will be performed with JL-Yellow.
June 19th, 2018
StrainSEEK V2s were loaded on MiSeqs for 2x150bp reads. A high quality run is underway.
June 20, 2018
Reads from 2 candidate Jamaican Lions captured by NEB Ultra FS kit and Agilent SureSelect and mapped with CLC v9.0.1 to canSat3 at 95% Length fraction ad 97% similarity fraction.
June 21, 2018
Raw sequencing reads from ILMN MiSeqs
DASH Blockchain Hash for RSP11075
DASH Blockchain Hash for RSP11076
More HMW weight DNA being prepped for Sequencing with Pacific Biosciences.
We gave a lab tour to Joshua from DASH today and were pleased to get some DASH Schwag.
June 22, 2018
After confirmation of Type II status with youPCR and Illumina sequencing, we are scaling up the DNA purification to get over 20ug of DNA (>40kb) for Pacific Biosciences sequencing. Robotic DNA preps and Manual DNA preps were performed to ensure high quantities of high quality DNA preparations.
Ampure is used as the final step after a chloroform extraction. For more information on the methods see the Medicinal Genomics MIP protocol.
Robotic DNA purification was performed on sterilized mature stalks. 100mg of stalks are cut into small pieces (<4mm) and placed in a SenSATIVAx Lysis buffer with 2.5ul of Proteinase K. 9 steel ball bearings are placed in each tube for 45 minutes vortexed lysis. SenSATIVAx is also used to cleanup this lysis.
June 23, 2018
Proverde Laboratories has developed methods that enable Chemical profiles from immature leaves. These are helpful to confirm genetic findings (Bt:Bd status) and provide terpene information. Initial analysis on two seedlings confirms Type II status and non Myrcene dominant terpene profile. Terpene profiles should be repeated with final flowering as literature exists that demonstrates terpene expression can change more readily over the course of the growth than cannabinoids content. Booth et al have an excellent paper on the the genetics of cannabis terpene expression. This work was presented by Dr. Page at CannMed 2017.
June 24, 2018
Grower reported that the plant is in vegetative growth phase and has been cloned out for distribution to various legal growers in the state to reduce single point of failure risks. We are having discussions with various parties about immortalizing this strain and offering DNA purification for other researchers that want access to fully sequenced cannabis gDNA. Live tissue can only be circulated in the state of Mass with proper permitting until further federal legalization occurs.
June 26, 2018
DNA was QC’d and prepared for overnight shipment to a Pacific Biosciences sequencing partner. Over 20ug of DNA was isolated.
June 27th, 2018
Pulse Field Gel Electrophoresis was performed by our collaborators to better assess the molecular weight of the DNA. Stem Preps have more fragmented DNA than the MGC MIP prep. Pacific Biosciences SMRTbell Libraries are being constructed with DNA from Lane 4. Alternative DNA purification techniques are being explored to deliver higher quantities and of HMW DNA to maximize the Read Length of Pacific Biosciences platform.
June 28, 2018
Multiple more attempts at uHMW DNA purification were performed. The following literature was helpful in the devising the approach. The Agilent gDNA tape station has limited resolution past 20Kb and alternative sizing platforms like SAGE Pippin Pulse power source and Advanced Analytical FEMPTO pulse need to be considered.
Modified Robotic Stem Preps are being considered to ensure lower Chloroplast DNA. Nuclei Preps that don’t require Liquid Nitrogen are also being investigated. A hybrid Qiagen + SPRI method was investigated yielding over 20ug of DNA. 200mg of leaf tissue is lysed and placed in a Qiagen shredder column. Potassium Acetate precipitation is performed followed by a SPRI of the resulting supernatant. DIN of 7.3 is the best quality yet.
June 29, 2018
SMRT Bell Library Prep was performed and Sized by our collaborators. Library is sufficient to run on a SMRT cell @ 29Kb but we should push for a 45-50kb given the newer sequencing chemistries. Sizing was performed on a SAGE Blue Pippin Prep and analyzed on an Advanced Analytical FEMPTO Pulse. 15Kb cut off was performed to deliver a 29Kb average size distribution with a short tail of molecules past 80Kb. There are
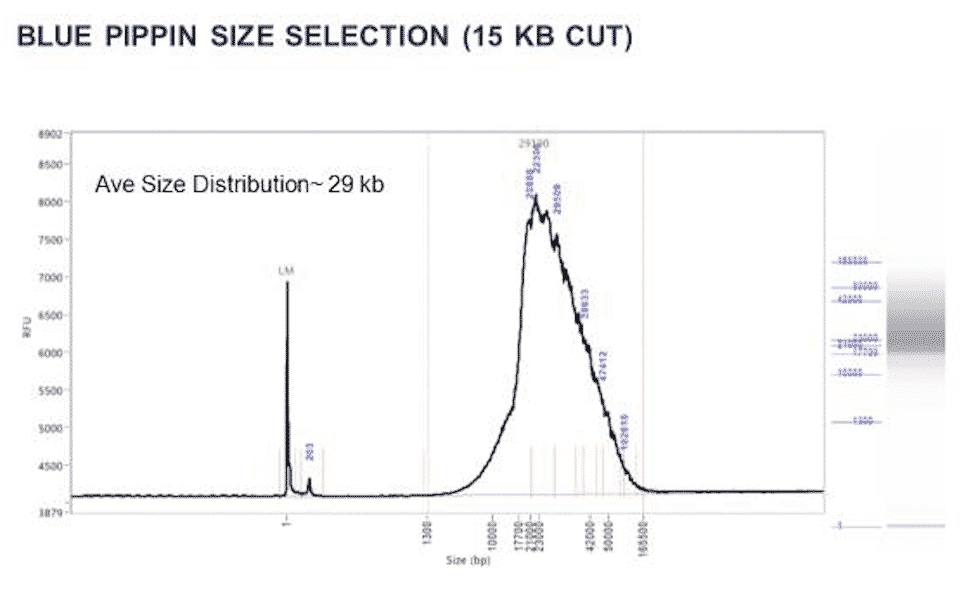
June 30,2018
We are preparing for shipment of DNA on Monday for 40kb + libraries.
July 2, 2018
Modified Qiagen DNA Purification
Plant leaves were placed in a -80°C freezer until sufficiently frozen. Once frozen, the leaves were ground to a fine powder, and 100 mg of ground material was aliquoted into 2 mL tubes. 800 uL of Qiagen Food Lysis Buffer (CTAB) was then pipetted onto the plant material, along with 4 uL RNase A. Six metal beads (Liam’s 02Jul2018 extraction did not have beads) were then added, and the tubes were vortexed. The tubes were then incubated at 65°C for 10 minutes, and were vortexed at the 2, 4, and 8 minutes. Following the incubation, the DNA purification was completed with the Qiagen DNeasy Plant Kit. 260 uL Buffer P3 was added to the lysate, vortexed for 5 seconds, and incubated on ice for 5 minutes. The lysate was then centrifuged for 5 minutes at 14000 rpm. Then, the lysate was pipetted into the QIA shredder Mini spin columns, placed in a 2 mL collection tube and centrifuged for 2 minutes at 14,000 rpm. 450 uL of the flow through was pipetted into a 1.5 mL microcentrifuge tube, and 675 uL of Qiagen Buffer AW1 was pipetted onto the flow through and immediately mixed. 650 uL of the mixture was then pipetted into the DNeasy Mini spin column, which was then placed in a 2 mL collection tube and centrifuged for 1 minute at 8500 rpm. The flow through was discarded and the remaining sample was pipetted onto the spin column and centrifuged at 8500 rpm for 1 minute. The collection tube was then replaced, and 500 uL of Buffer AW2 was added to the spin column and centrifuged for 1 minute at 8500 rpm. The flow through was discarded, and 500 uL of AW2 was again added and centrifuged for 2 minutes at 14000 rpm. The flow through was discarded, 500 uL of AW2 was again added and centrifuged at 14000 rpm for 2 minutes. Lastly, the DNeasy column was placed in a 1.5 mL microcentrifuge tube, 100 uL of Buffer AE was pipetted directly onto the membrane, was incubated at room temperature for 5 minutes, then was centrifuged at 8500 rpm for 1 minute to elute the DNA. The DNA was then quantified with a Qubit to determine concentration and run on an Agilent 4200 tape station to determine molecular weight.
DNA shipped to collaborator for PFGE.
July 5, 2018
Initial BLASR analysis from a Sequel test chip is pointing to %4.7 – %8.8 Chloroplast DNA. This is lower than expected as we did not use a pure nuclei prep. nuclei_preps
July 6, 2018
Cannabinoid and terpene profiles were analyzed at Proverde Labs demonstrating Type II status where Myrcene is not (yet) the dominant terpene. Terpenoid content can change as the plant moves into flowering.
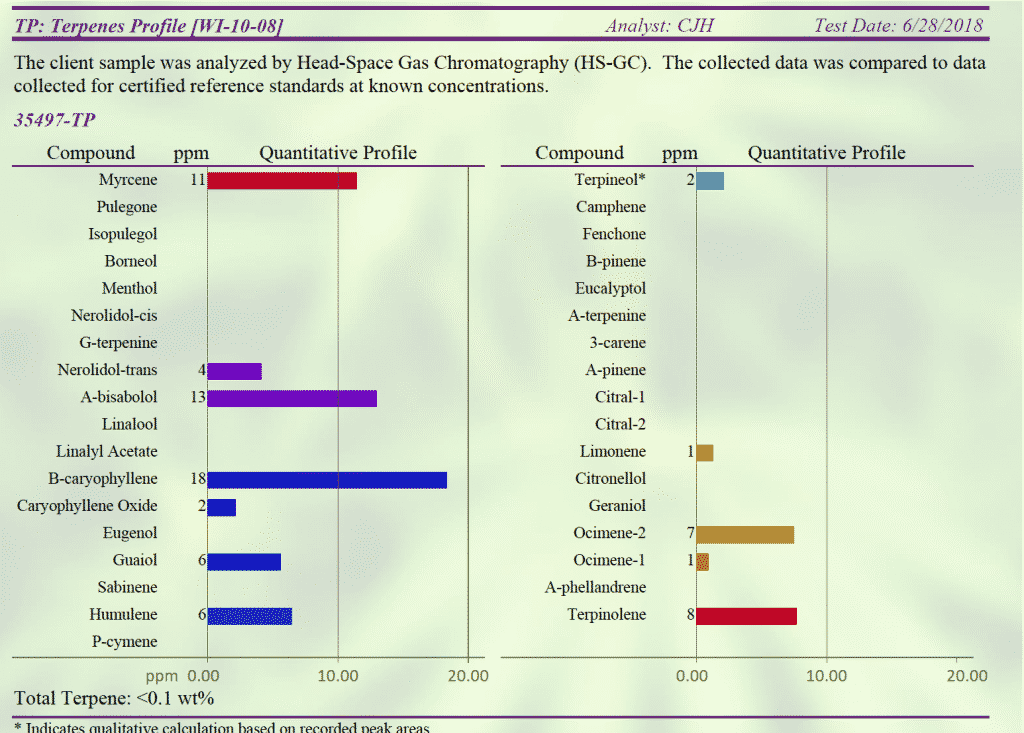
jamaican_lion_yellow_cannab_terps
July 7, 2018
PFGE of the Modified Qiagen Prep. The 65C Vortex step in hindsight is probably not a good idea. More DNA prep conditions will be explored.
June 28th SMRTbell library generated 7.26Gb of Sequence with 30Kb N50s. Another Size Selection of this library has produced a 42Kb library size. Multiple chips will be sequenced with this library.
Depiction of the evolution of Pacific Biosciences SMRT cell technology. The Sequel Chip that performed a 10X Cannabis Genome in a single run is on the right.
July 9th, 2018
Further optimizations on DNA purification are ongoing to push the DNA size out to 50kb+. The Agilent platform is a bit limited in guiding this approach so we are reaching out to collaborators to rent their gear to guide this optimization. This should save on CapEx purchasing of Pulse Field Gel Electrophoresis equipment. Many thanks to our collaborators for helping us on this front.
July 10th, 11th 2018
July 12th, 13th, 2018
July 19th
Multiple SMRTCell runs have generated 91Gb of sequence. Assuming a 1Gb genome, this is 91X coverage where 50% of the bases are in reads over 32Kb. This should substantially improve the reference genome. Reads are now in DNA Assembly. This step can takes days to weeks to perform on high performance computers. More DNA is being purified and sized for attempts at longer libraries.
July 28th, 2018
Helpful Talks Regarding DNA Assembly.
-Sarah Kingan: https://www.youtube.com/watch?v=SKL1y8klKUM
-Adam Phillippy: https://www.youtube.com/watch?v=zGYbiArVMV0
-FALCON: http://pb-falcon.readthedocs.io/
We now have a FALCON assembly. The Cannabis Genome is now in better shape than the Human Genome Project during its famous 2001 Whitehouse celebration. No other Cannabis reference to date meets this criteria (greater than 500kb N50). THCAS and CBDAS are now on the same contig for the first time in history and the genome size is greater than a Gigabase. The assembly is in the polishing steps and will be uploaded in a few weeks once some error correction and QC is performed to screen for microbial contaminants etc.
July 30th, 2018
Assembly statistics were calculated with Quast.
The N50s are improved by over 200 fold from assemblies performed in 2011. More recent long read assemblies are 4-12 fold less contiguous than the DASH funded Jamaican Lion. This is likely the result of longer reads from higher molecular weight DNA and deeper coverage. THCAS and CBDAS are finally on the same 411kb contig. OLS and PT are on contigs close to a Megabase in size.
Here is the DASH blockchain information for the gz file:
filename: JamaicanLion_FALCON_primary_contigs_unpolished.fa.gz
SHA256: 1f6dc3d2c1f69428e67b8f9b7712a94224467cd5744b481ffb5a41f8d12a3cbe
TransactionID: 9622725bc9093625c24bac7574b96196fb579dc49f9803b70dbae18f337f731c
FASTA file will be loaded to this site once a preliminary QC is complete. Note- this is unpolished data. There will be higher indel error until Polishing is complete.
August 1, 2018
Mapping ILMN StrainSEEK (3.2Mb target) reads to the PacBio Falcon Assembly and the CanSat3 reference.
CanSat3 reference maps 2.131M reads while the PacBio reference maps 50% more reads (3M). More reads are mapped as pairs with the PacBio assembly.
Better mapping rates despite an unpolished reference is impressive but we still need to polish the PacBio data as we can see some indels in the THCAS sequence.
While some indels still exist in the PacBio data, polishing steps should correct these. We can also see that the long read assembly is now resolving the collapsed THCAS pseudogene cluster. The Top track is the CanSat3 assembly and the bottom track is the PacBio assembly. One can see multiple single ended reads (red and green and some paired reads that are mis-mapped onto THCAS and presumable have better places to map in the more complete PacBio assembly.
August 3rd, 2018
BLAST of contig000692F (CBDAS and THCAS contig) demonstrates multiple copies of synthase genes.
August 4, 2018
The genome is now on the IPFS.
August 6th 2018
Quast was utilized to assess the assembly quality
August 7, 2018
Efforts to obtain 50Kb+ DNA isolations are under way using a hybrid method from protocols.io and Circulomics
high-molecular-weight-gdna-extraction-after-mayjon-khkct4w
sigmainvoicecc_mckernan-545450854
DNA preps are going onto SAGE 30Kb PacBio gel cassettes for sizing 35Kb-80Kb.
August 11, 2018
In order to assess genome completion we have downloaded an installed BUSCO.
Assessing genome assembly and annotation completeness with
Benchmarking Universal Single-Copy Orthologs
Duplicated and Fragmented genes are expected with an unpolished reference.
August 15, 2018
Our SAGE Pippin Prep had a current leak. We have swapped out the instrument ($1500) and should have HMW DNA through size selection this week.
We also received a call from Canada Cannabis Association who has taken an interest in this project as a method to protest these patents in Canada. The patents have issued in Canada on August 14th with reduced claim scope.
Protest from Cannabis Canada Association-
http://brevets-patents.ic.gc.ca/opic-cipo/cpd/eng/patent/2911168/images.html?page=1&frenchDocType=Poursuite-Amendment&englishDocType=Prosecution-Amendment&modificationDate=20171225&scale=25&rotation=0&type=&objectName=A1001001A18G27B04404I85157&numPages=9&query=*&start=301&num=50
Response from “inventors”
http://brevets-patents.ic.gc.ca/opic-cipo/cpd/eng/patent/2911168/images.html?page=7&frenchDocType=Poursuite-Amendment&englishDocType=Prosecution-Amendment&modificationDate=20171231&scale=25&rotation=0&type=&objectName=A1001001A18H02B04500H34625&numPages=13&query=*&start=301&num=50
Here is the complete file wrapper-
http://brevets-patents.ic.gc.ca/opic-cipo/cpd/eng/patent/2911168/summary.html?query=*&start=301&num=50&type=
August 16, 2018
Aiming for 50kb+ libraries. Size Selections with SAGE Pippin Prep looks cleaner with higher DIN scores. Running on a Fempto Pulse next.
Our Collaborators at NEB ran a Fempto Pulse check on the latest prep. It looks like it needs a size selection to improve upon the 32Kb library.
August 17, 2018
Assembly Polishing by Arrow significantly improves the assembly.
7 years ago today we uploaded the first Cannabis genome to the Amazon Cloud.
Today we have a Polished Pacific Biosciences genome reference that is 303X more contiguous. This reference has been loaded up to Mega.nz and to IPFS.
The BUSCO results are significantly improved after polishing showcasing 92.9% of the genes being intact.
The Polishing step is known to remove many indel (insertion/deletion) errors in the assembly. Indels cause frameshift mutations in genes and this significantly lowers the BUSCO calculations of Complete genes. Below is a region of THCAS sequenced with a SureSelect exome capture on an ILMN miseq. One can see the alignments to canSat3 scaffold19603 (THCAS) and Jamaican Lion contig 000692 (THCAS). A deletion of a single A in the PacBio Jamaican Lion reference would cause this single exon gene to be frame shifted and likely non-functional. The program Arrow combs through all of the quality files in the PacBio movies to correct these artifacts. The polished version of the assembly is seen on the bottom depiction.
Quast report on the Polished Assembly.
August 20,2018
More analysis work is being performed to assess the completeness of the genome.
1)Run BUSCO on the genome plus the haplotigs. This improves the results to 93.6%.
August 21, 2018
Map Whole Genome Shotgun ILMN reads to the genome to assess read mapping rates. Over 96% mapping.
Run SAGE pippin prep to size cut the latest Fempto DNA. 50Kb-80kb size cut resulted in no DNA.
August 23, 2018
Upload the genome to NCBI.
August 24th, 2018
Map 6 other Whole Genome Shotgun projects to the new reference.
Re-Extract DNA from young leaves and hydro-roots.
The long RNAse step is likely enabling other nucleases to digest the DNA. Replace it with a Proteinase K step in the CTAB extraction.
Double the Proteinase K amount. Move it to the front of the prep so as to digest nucleases as soon as possible. Put the RNAse at the end of the prep.
Highest DIN scores yet. Beads looked viscous and clumped. 40ng/ul. Drive DNA to NEB for Fempto QC
August 25, 2018
Run MUMmer on Jamaican Lion (QRY on X axis) x Cannatonic (REF on Y axis).
Jamaican Lion is nearly 2X larger than the Cannatonic assembly. The white spaces on this dot plot is all new genomic sequence.
97.7% of the Cannatonic assembly can be found in Jamaican Lion and 3.3M SNPs.
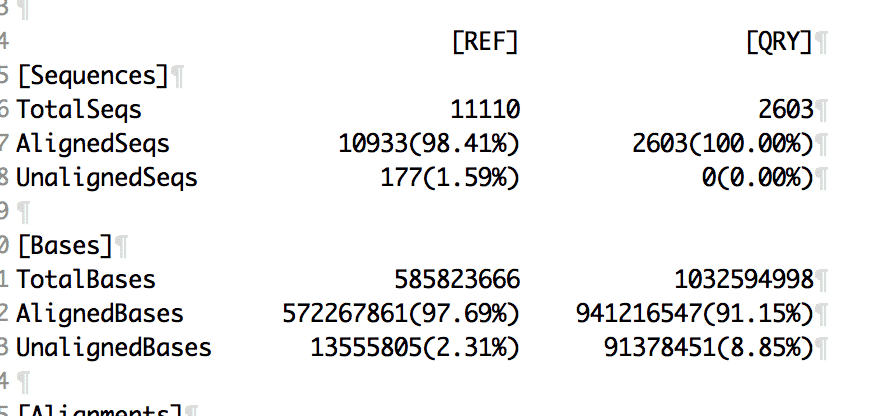
August 27th to August 31st.
Cannabis Science Conference in Portland OR. Jamaican Lion still appears to be the most contiguous public assembly. SunRise genetics presented a very nice but a non-public assembly with N50s in the 750kb range. The Hi-C data below is pushing the Jamaican Lion assembly over 1.858Mb N50s.
Phase Genomics libraries passed QC and began sequencing.
10M paired reads were run on an ILMN Miseq to QC the library before 150M more reads are run.
The Phase Genomics HI-C scaffolded assembly can be downloaded below. If you sign up with the email address you will get periodic updates on the assembly status.
https://mega.nz/#!gdB0WSJS!Cdit3Ey7a1ocsh8QJOTM-PHaRIdGgs9NAjZ6SHPzBaA
N50s are now at 1.858Mb
BUSCO analysis was completed on the other relevant genomes.
September 4th- 9th, 2018
Additional HI-C reads were generated pushing the assembly up to N50s of 2.57Mb.
Latest Assembly
https://mega.nz/#!RZAAQY6B!BlG2qvB7-xk-VIYaJOTQ-SSEAK0diJ64gTRMW85_5Ls
September 10th- September 13th, 2018
Nuclei Preps were prepared with Agarose plugs. This protocol has a 2 days gentle lysis protocol once nuclei are captured in plugs. Very High DIN numbers. Need to run on Fempto to confirm. 40ng/ul. -80C Mortar and Pestle results most remarkable.
September 17th – September 21, 2018
Fempto Pulse of -80C Mortar & Pestle + Agarose Plug Nuclei Preps
DNA shipped to service provider.
Luca Massimino Aligned all of the RNA-Seq data from van Bakel et al. to the Jamaican Lion Assembly.
65M HiC reads are beginning to gel into chromosomes with Juicer/Juicebox.
September 21-September 28, 2018
Over 600M HiC reads were sequenced.
338M reads passed Phase genomics QC.
mckernan_jamaican_lion_bam_to_mate_hist_qc_report
Mapping with the Arima pipeline and SALSA delivers a 5.4Mb N50 genome with under 900 contigs.
HI-C data is highlighting some potential mis-assemblies.
October 1 – October 7, 2018
Install Polar_Star to look for coverage deviation that might signify assembly misjoins
Install Purge_haplotigs to look for polymorphic haplotigs that might be ballooning the assembly length.
Install Bandage to look at Graph files from the assembly.
Hi-C data highlights some potential assembly errors. Polar Star and Purge Haplotigs will be run to clean this up.
October 8th- October 14, 2018
Graph file from Bandage.
Purge Haplotigs was run to produce a haploid only reference.
Max Press
Ran Proximo on the data. 10 Chromosomes are beginning to emerge.
October 15- October 22, 2018
Longer Pac Bio Library looks good.
New Polished Assembly with 34Gb of V6 chemistry. 125Gb total.
==================== Scaffolds | withGaps | withoutGaps ==================== #Seqs | 558 Min | 32,282 1st Qu.| 363,087 Median | 953,411 Mean | 1,924,832 3rd Qu.| 2,287,198 Max | 32,623,077 Total | 1,074,056,627 n50 | 3,811,003 n90 | 929,797 n95 | 568,387 ==================== Contigs | withNs | withoutNs ==================== #Seqs | 558 Min | 32,282 1st Qu.| 363,087 Median | 953,411 Mean | 1,924,832 3rd Qu.| 2,287,198 Max | 32,623,077 Total | 1,074,056,627 n50 | 3,811,003 n90 | 929,797 n95 | 568,387 ==================== No Gaps! ==================== Non-gapped Ns Count: 0 Alt Contigs- https://mega.nz/#!xQRBXIZS!02qfzgofFgyNIMdKXDAwteqEuX137KrlH4Gtm_FM7Hs Primary Contigs- https://mega.nz/#!QYAGkaSA!Jvse2Lk5jQsh290ALRIaXzfQNIEhNgbu1yVfUrg1i9c
CannMed 2018 Presentation
kevin_main_update_cannmed_2018
Dash Hash
013a5260645593dc33423c2808f2a5b892b220359d5f9076c67be01ed8b41d39
V6 Falcon Unzip Primary Assembly. 3.79Mb N50.
https://mega.nz/#!QYAGkaSA!Jvse2Lk5jQsh290ALRIaXzfQNIEhNgbu1yVfUrg1i9c
97cc128b8e7ba2cbb40bc18e1103250201e80e904bba8c04e204a1cf96a6418c
Haplotig AltContigs
https://mega.nz/#!xQRBXIZS!02qfzgofFgyNIMdKXDAwteqEuX137KrlH4Gtm_FM7Hs
a9eb9883181f218ef0b106999806889cc30d31c356e985012e74aaf7eb99c5d2
©2025 MEDICINAL GENOMICS CORPS
Copyright all right reserved.
Signup to receive email updates on blog posts, podcasts, webinars, new products, and more!
Join us at the CannMed 25 Innovation and Investment Summit, an exclusive experience of discovery, development, and networking with innovators from around the world.