Our latest pre-print study, provides evidence that two competitors’ Aspergillus qPCR assays inappropriately amplify Aspergillus tamarii, causing false-positive results that could potentially lead to millions of dollars in crop loss.
Investigating Discordant Results
A customer recently contacted us about discordant results for some flower samples between our PathoSEEK 5-Color Aspergillus Detection Assay and our competitors’ assays. More specifically, a grower failed for Aspergillus at a lab using one of our competitor’s assays. They then contacted a lab using PathoSEEK, and passed. With hundreds of pounds of cannabis on the line, they needed to know which result was right.
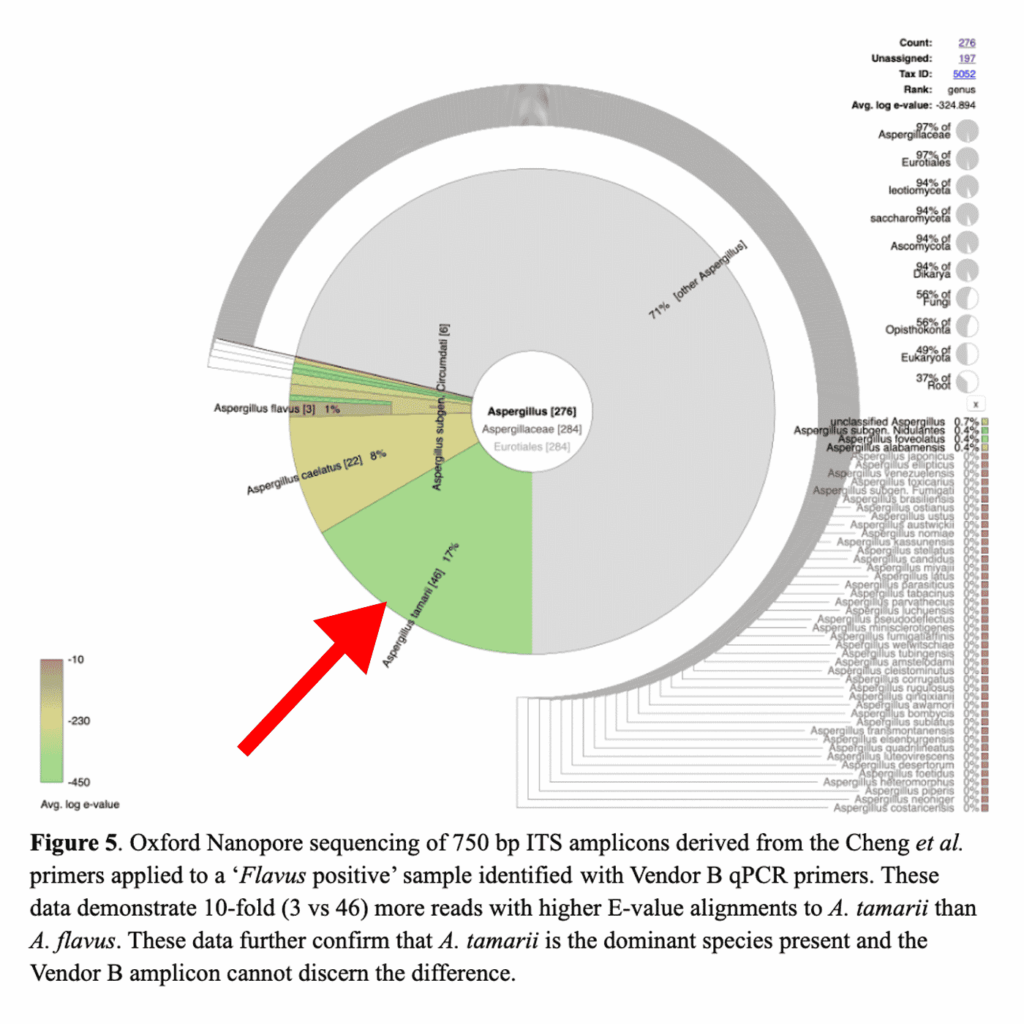
To understand this discordance, we first sequenced the microbiome of the samples in question using our PathoSEEK ID Service. Interestingly, we found the dominant species to be Aspergillus tamarii, a non-pathogenic species of Aspergillus that is not included in this state’s testing regulations (see the red arrow in the figure below).
You may notice that the plot includes a small percentage of Aspergillus flavus; however, that does not mean A. flavus was present in the sample. The plot is not showing a breakdown of all the species present on the sample. The plot is showing the software’s attempts to map DNA fragments to known Aspergillus species. You will see that only 3 of those fragments were mapped to A. flavus, and the red coloring of the pie slice indicates the software is not confident in that match. On the other hand, the software feels very confident in the 46 reads it mapped to A. tamarii. The green coloring indicates a low E-value, which means there is very little doubt the DNA could be anything but A. tamarii.
We knew A. tamarii wouldn’t trigger a positive result on our PathoSEEK assay because we tested the organism as part of the inclusion/exclusion requirement for our AOAC PTM report.
When we reviewed our competitors’ PTM reports, we saw that A. tamarii is not in their inclusion/exclusion table (Competitor A, Competitor B). However, after digging a little deeper we found a scientific poster that Competitor A published, which states their assay hits several off-target species of Aspergillus, including A. tamarii (see below)

Testing A. tamarii on Competitors’ Assays
This led us to suspect that our competitors’ assays were unable to differentiate A. tamarii from A. flavus. We sourced live A. tamarii organisms from ATCC and with the help of independent cannabis testing labs, tested it on our competitors’ assays, using the current protocols as of September 12, 2022. As we suspected, both assays returned positive results.
Evaluating qPCR Primers
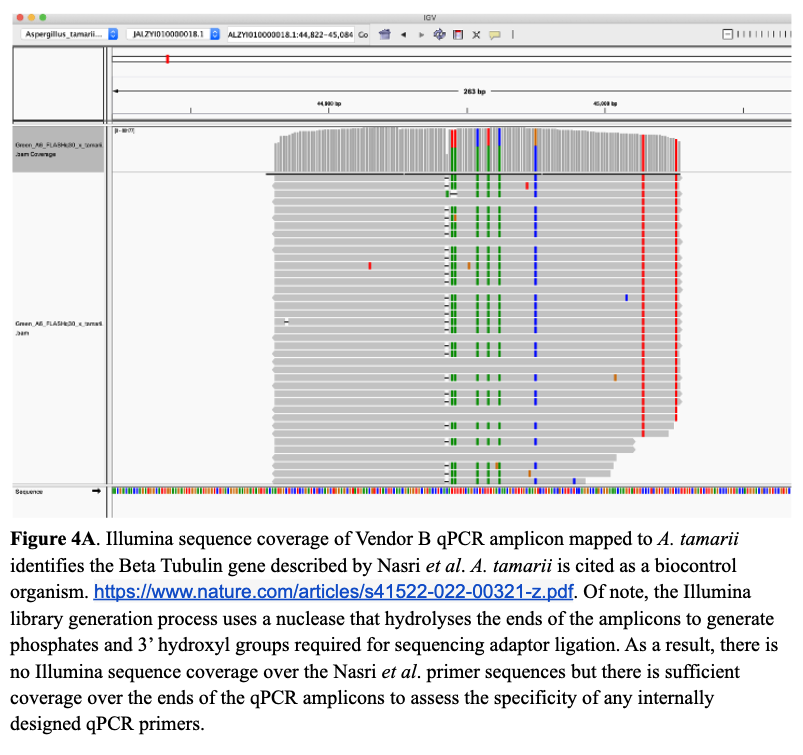
Next, we wanted to understand why A. tamarii caused problems with our competitors’ assays. Without access to their primer sequences, the next best thing was to sequence the amplicons. Doing so revealed that both competitors target the Beta Tubulin gene described by Nasri et al. Unfortunately, the Beta Tubulin gene is not specific to the four pathogenic species of Aspergillus (A. flavus, A. fumigatus, A. niger, and A. terreus), and therefore, the amplicons map well to A. tamarii.
Conclusions
We believe the results of this study explain the discordant results between our 5-Color Aspergillus Detection Assay and our competitors. Although one may look at the results from three competing assays and assume the two that agree are correct, that is not the case here. We have shared this information with customers and many state regulators that require Aspergillus testing, so they understand the discordance.
These findings may also have major implications for cannabis growers whose livelihoods depend on clean microbial testing results. False positives can potentially cost them millions of dollars and force them to unnecessarily irradiate their crops, which compromises quality. In some states, a positive Aspergillus pathogen results in the destruction of the crop. Many people have voiced that false positives are acceptable, but false negatives are a patient risk. We prefer to have neither as false positives elevate the price of legal cannabis and drive consumers back to untested black markets.
Growers should contact their testing lab to make sure they are using PathoSEEK to test for Aspergillus. Otherwise, they are at risk of failing for an off-target organism.
We acknowledge that this study is not peer-reviewed. While peer review is valuable, so are timely, actionable results. These findings are easy for labs to verify for themselves by simply ordering ATCC1005 and running their own experiments. And we encourage them to do so! The community can quickly assess the veracity of this claim before peer review even gets its boots on.
More cannabis microbiome surveys are needed to better understand how prevalent A. tamarii is in the cannabis field. In addition to the customer’s samples that triggered this investigation, we analyzed the cannabis microbiomes included in the Comeau, et al. study and found that 1 in 50 had high percentage hits to A.tamarii.
We are happy to collaborate with growers, labs, and/or regulators looking to investigate these discordant results.
Read the full pre-print study at https://zenodo.org/record/7071829#.Yzw1guzMJqu








