A new collaborative study (preprint) consisting of scientists from Medicinal Genomics, The University of Florida, Pacific Biosciences, and Minnibis, is opening our understanding of the cannabis genome. It has gotten to a point where breeders can use genomic data to make informed breeding decisions about cannabinoid production, pathogen resistance, and more. These public data will drive a genomic revolution in cannabis cultivation.
How we got here
In 2018, Medicinal Genomics began a project to create the most complete cannabis reference genome. The team “shotgun sequenced and assembled a Cannabis trio of the Jamaican Lion cultivar (sibling pair and their offspring)” using the Pacific Bioscience Sequel II platform. This resulted in the most contiguous Cannabis sativa assemblies to date.
Next, the team performed full-length male and female mRNA sequencing to identify the regions of the genome that encode the proteins the plant uses to carry out its functions. Finally, the team mapped whole-genome sequence data from a diverse set of 42 male, female, and monoecious cannabis and hemp varietals to the reference. This allowed them to identify high-impact variants that may disrupt gene function and evaluate copy number variations (CNV).
CNV maps like the ones shown below may play an important role in breeding cannabis and hemp varieties for cannabinoids, terpenes, and pathogen resistance. Cultivators and breeders should consider whole-genome sequencing in order to make informed decisions that can accelerate their efforts. Medicinal Genomics can help, contact us for more information.
Cannabinoid synthase genes
Using the coverage maps across THCA synthase (THCAS), cannabidiolic acid synthase (CBDAS) and cannabichromenic acid synthase (CBCAS), cannabis plants can be classified based on primary cannabinoid expression–Type I, II, and III plants.
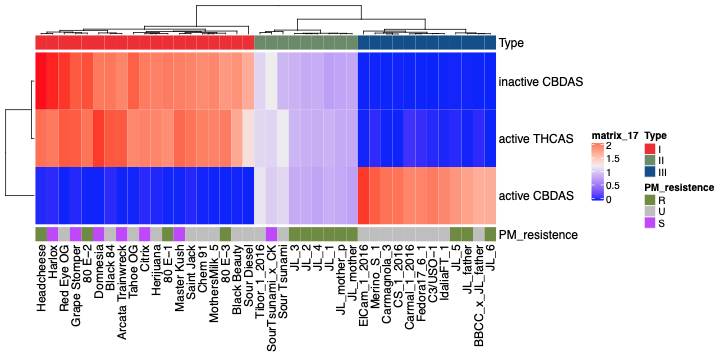
The preprint study states, “Plants lacking a functional CBDAS gene are Type I plants. Type II plants have both functional genes and synthesize both THCA and CBDA. Plants with no functional THCAS gene and a functional CBDAS gene are Type III plants. Plants lacking both functional genes are Type IV plants and only synthesize the precursor cannabigerolic acid (CBGA). While deletions of entire THCAS and CBDAS genes are the most common Bt:Bd alleles observed, it is possible to have plants with these genes where functional expression of the enzyme is disrupted by deactivating point mutations.”
In all, the team evaluated 39 cannabinoid synthase genes for CNV across 40 genomes. Of interest is the frequent deletion of the CBCAS gene cassette (highlighted in red), which contains 8 nearly-identical genes.
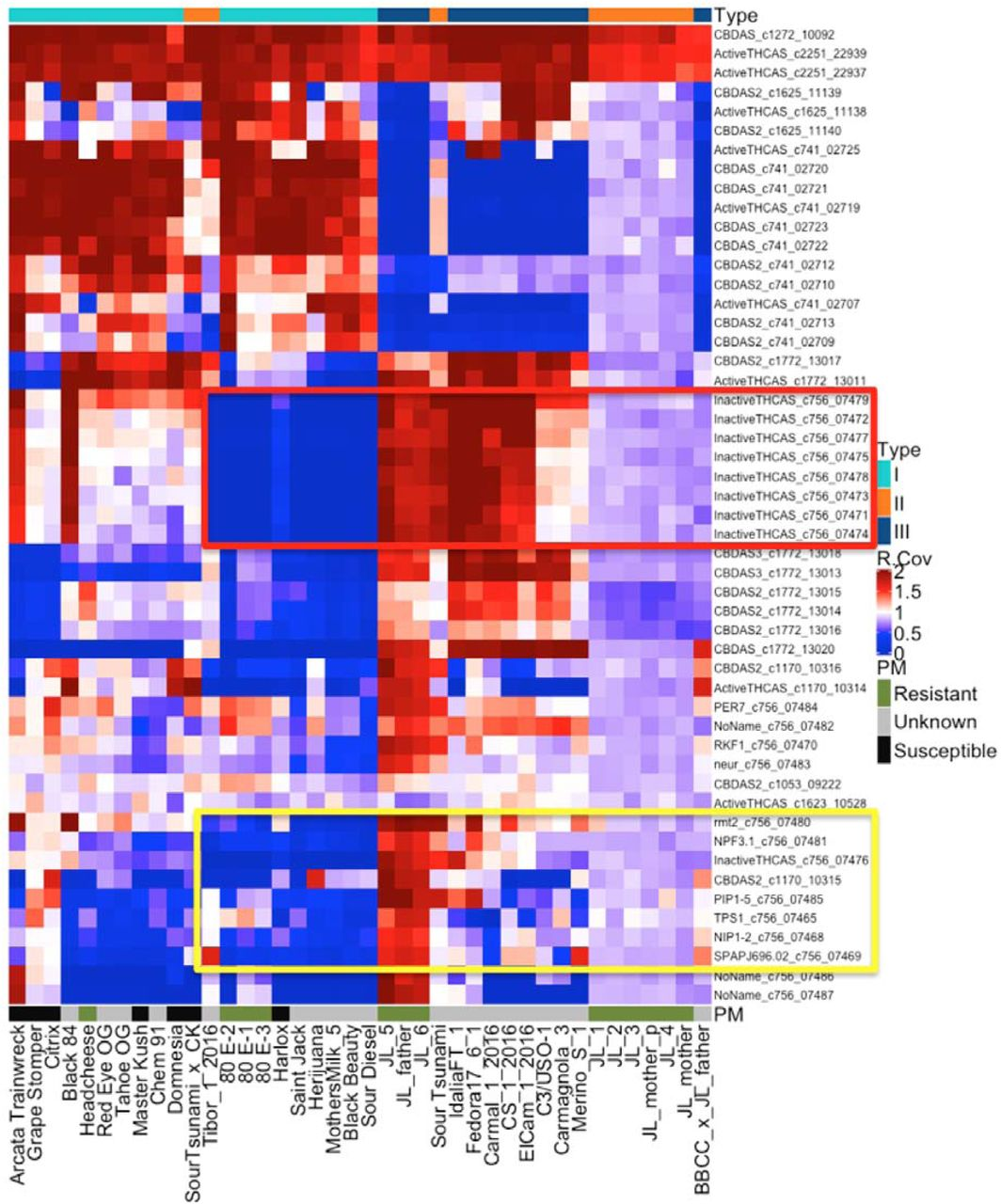
This observation could be a clue to understanding why some Type III plants still produce small concentrations of THCA. The authors of the preprint article write:
“Winnicki et al. demonstrated that multiple cannabinoids can be expressed from a single cloned synthase gene by modulating the yeast growth conditions (US patent 9,526,715 & 9,394,510)(Peet 2016) (Winnicki 2016). Hemp lines have also been more difficult to grow while maintaining a THCA concentration below the 0.3% THCA limit mandated in many jurisdictions. In particular, the THCA levels appear to increase in varieties from equatorial climates (Clarke 1996). Thus, it is possible that the presence of this cassette or other cannabinoid synthase CNVs are responsible for low levels of promiscuous THCA expression in some plants lacking a THCAS gene (Type III plants).”
This CBCA deletion also contains several pathogen response genes (highlighted in yellow), which could mean breeding for low THCA could also select for pathogen susceptibility. Cannabinoid synthase CNV maps could play an important role helping breeders create compliant, pathogen-free hemp cultivars that do not synthesize residual THCA.
Pathogen response genes
Speaking of pathogen response genes, so far the team has identified 82 genes that may play a role pathogen resistance.
They analyzed the data from the 42 genomes for copy number gains and losses among these genes and noticed PM-susceptible cultivars had deletions of a thaumatin-like protein (TLP) gene, while PM-resistant cultivars contain copy number gains in the same gene. The team also observed cultivars with endochitinase CH25 and lack of mildew resistance loci O (MLO) correlated with resistance to PM.
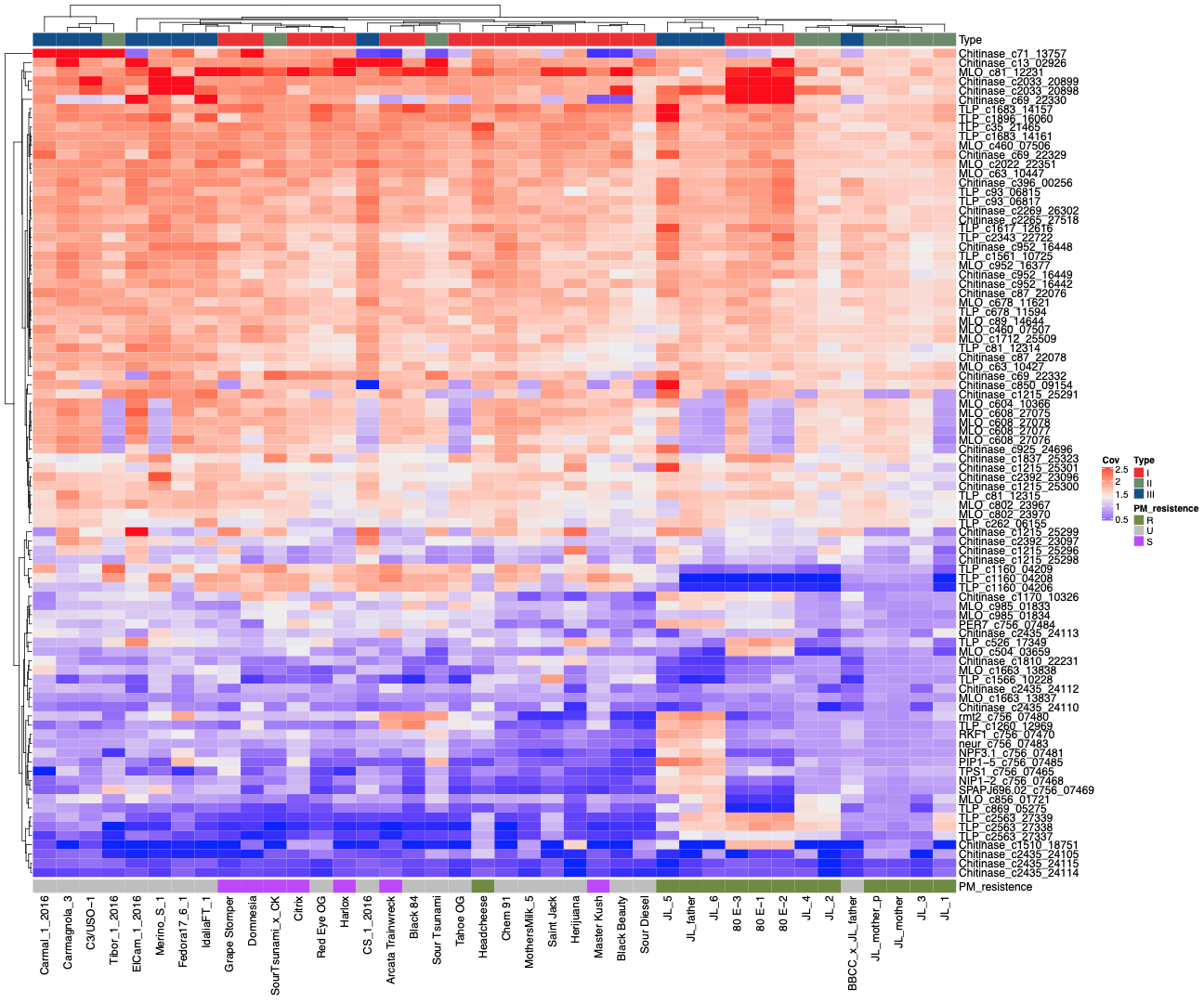
Many cannabis plants are believed to be powdery mildew-resistant, but the genetics of this allele have yet to be identified. Identification of the PM-resistance marker can lead to more targeted breeding for healthier plants. Furthermore, the authors speculate that “cloning and expression of the genes in a non-pathogenic bacterium could even enable the development of foliar enzymatic sprays against epiphytic pathogens such as PM.”
However, if you look at cannabis’ closest relative, hops, you will see it has a complex pathogen response against PM that involves multiple genes. So it is unlikely that a single gene is responsible for PM resistance in cannabis.
Further functional studies need to be performed before definitive markers can be established, but breeders and cultivators can benefit from sequencing their cultivars to look for trends among these genes and breed accordingly.
Other insights
The study includes other interesting insights into the cannabis genome, including sex determination, terpene synthase, and yield-related genes. Be sure to read the pre-print for more information
Sequence now
Cultivators and breeders looking to create novel varieties, select for pathogen resistance or stabilize lines should consider whole-genome sequencing in order to make informed decisions that can accelerate their efforts. Medicinal Genomics can help, contact us for more information.